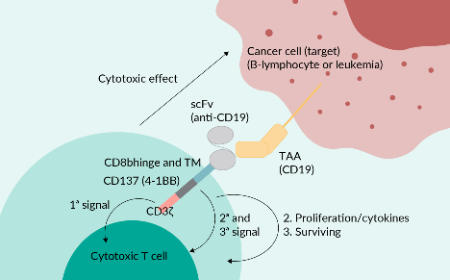
Cell & Gene Therapy Insights is an online, open-access, peer-reviewed journal dedicated to the interdisciplinary exploration and advancement of cell and gene therapy. With a translational focus, we connect innovative research to practical clinical applications, providing valuable insights into one of biotechnology’s fastest-evolving fields.
We address the key challenges and latest developments across advanced therapies, publishing original research articles, expert reviews, commentary, clinical trial reports, and more. Visit the Cell & Gene Therapy Insights Journal page for our complete collection.
Our popular webinar series provides expert-led discussions on important developments and methodologies in the field, designed to support ongoing professional growth.
Explore our specialised channels, including detailed coverage of critical areas like the cell and gene supply chain, for targeted insights into manufacturing, logistics, and regulatory frameworks essential to therapy commercialisation.
Learn more about our journal’s mission and publishing criteria by visiting our aims and scope.
Sign-up for free to gain unlimited access to our extensive library of articles, webinars, podcasts, news, and interviews, and stay at the forefront of cell and gene therapy innovation.
If you’re interested in working with us, from sponsoring articles to webinars and more, view the media kit to find out more.
December 2025
Upcoming webinars

Keys to successful lentiviral vector scale-up
Uncovering CAR-T and CAR-NK cell potency using multi-analyte profiling
Automating and scaling T cell manufacturing in hollow-fiber bioreactors

Developing streamlined, flexible cell therapy process workflows: from upstream to formulation and fill-finish operations

Genomic integrity and safety assessment strategies for CRISPR-edited cell therapies
Defining effective antitumor T cell responses to guide next-generation adoptive cell therapies for solid tumors
Latest Articles

Advancing cell dissociation in bioprocessing: a novel, high purity trypsin-like enzyme for GMP workflows
Industry Insights: From rare disease to chronic indications, cell and gene therapies broaden their impact
Translating both a cure and a commercialization pathway: insights into a breakthrough gene therapy company’s journey
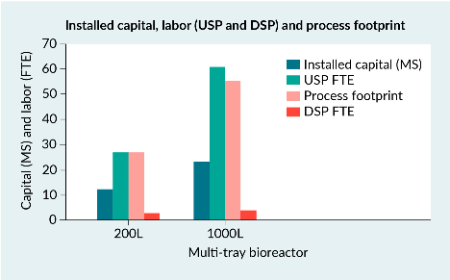
Scaling AAV5 production from bench to industrial scale: strategies for efficient, de-risked manufacturing
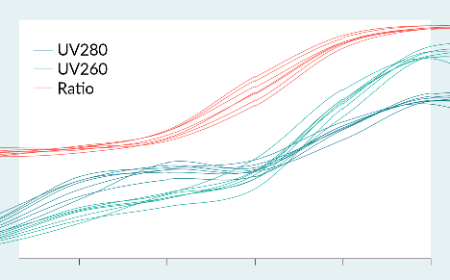
Accelerating AAV process development with high-throughput strategies

The next frontier of in vivo CAR-T: a conversation with Shon Green

Leaping forward: how China’s biotech evolution is capturing the in vivo CAR-T frontier

Scaling smarter: a case study in overcoming cell therapy manufacturing and regulatory hurdles