The progression & delivery of adoptive cellular immune therapies: leveraging single-cell resolution & novel algorithms to overcome the challenges associated with allogeneic & autologous immune cell therapies
Cell & Gene Therapy Insights 2020; 6(9), 1411–1430
10.18609/cgti.2020.155
The success and progression of chimeric antigen receptor (CAR) T cells in adoptive cellular therapies for B cell malignancies can be attributed to the effective engraftment, efficient expansion, and the persistence of the cells after transplant. However, that success has yet to be translated into solid tumors, which present their own set of distinct challenges. As the knowledge in the field grows and more data reach the public domain, it has become clear that both intrinsic and extrinsic mechanisms of tumor resistance need to be addressed in order to improve immune-based adoptive cell therapies (ACT). Computational approaches and novel algorithms are well placed to identify new genes needed to overcome resistance and enhance efficacy of the current and future ACT. This review discusses the current challenges for autologous and allogeneic ACT and how big dataset analysis is opening paths to overcome resistance and enhance the efficacy of ACT.
Introduction
The idea that immune ACT could play a central role in the fight against cancer was developed in 1964 by Alexander and Delorme [1]Delorme EJ, Alenxander P. Treatment of Primary Fibrosarcoma in the Rat with Immune Lymphocyte. Lancet 1964; 2: 117–20.. Their work demonstrated that sarcomas in rats can be treated by ACT of lymphocytes from immunized syngeneic animals. So far, only three immune ACT, using CAR T cells, are available on the market for the treatment of hematological blood malignancies, but the field is rapidly expanding. Currently, several clinical trials are running or approved, for both autologous and allogeneic immune ACT. This is due to incremental knowledge acquired in several fields, including stem cell transplantation, monoclonal antibody, and HIV research, together with the technical advances achieved in molecular biology. However, alongside the excitement of developing new therapies, the industry is faced with new challenges at the scientific, manufacturing, and regulatory level.
CAR T cell therapy uses autologous T cells isolated from patients, which are genetically modified to insert the CAR construct (Figure 1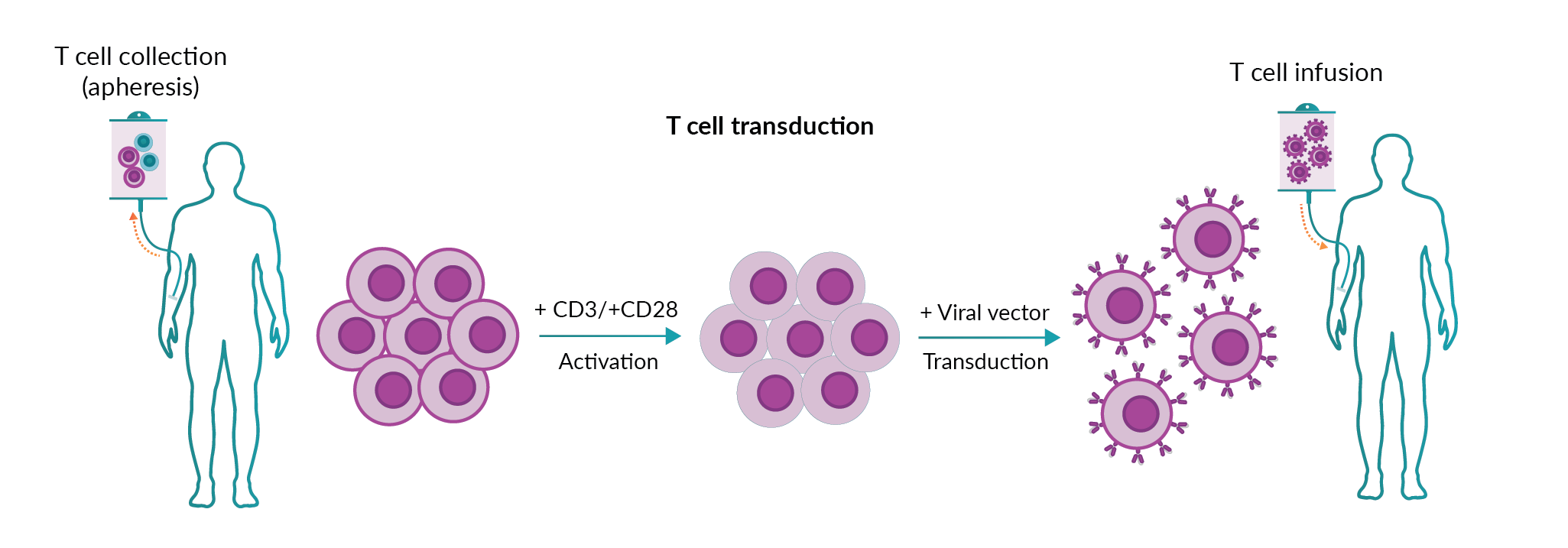
Despite these successes, almost 30% of patients with DLBCL are not able to receive autologous CAR T cell therapy due to low product quality during the processing and manufacturing of the CAR T cells [4]Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48., [5]Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.. This is mainly due to a reduction in T cell numbers or T cell exhaustion (lack of functionality) because of the severity of the disease and/or treatment [7]Majzner RG, Mackall CL. Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med. 2019; 25: 1341–55.. An allogeneic ACT could potentially address these issues, by providing an ‘off the shelf’ and reproducible alternative. An allogeneic product could be immediately available to the patient, reducing treatment lead time, and ensuring availability if redosing is necessary. Despite the promise, allogeneic ACT will face two major barriers to success; graft versus host disease (GvHD, whereby donor T cells attack the recipient cells), and rejection. Nonetheless, there are several approaches that different groups have developed to tackle these issues (reviewed in Depil et al. [8]Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.).
Next-generation sequencing and high throughput data approaches will play a key role in the identification of gene regulators or small molecules that can prolong the effectiveness of CAR T cells. For example, datasets such as the FANTOM5 consortium data have been employed in new approaches by Mogrify® to identify the optimal combination of transcription factors (TFs) required to directly convert any human cell type into any other human cell type [9]Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.. In addition, the same dataset has made it possible to identify enhancers and promoters that are important in T cell and macrophage differentiation by profiling human T cells [10]Schmidl C, Hansmann L, Lassmann T et al. The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood 2014; 123: 68–78.Schmidl C, Hansmann L, Lassmann T et al. The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood 2014; 123: 68–78. and monocytes [11]Schmidl C, Renner K, Peter K et al. Transcription and enhancer profiling in human monocyte subsets. Blood 2014; 123: 90–9.Schmidl C, Renner K, Peter K et al. Transcription and enhancer profiling in human monocyte subsets. Blood 2014; 123: 90–9.. This type of large-scale data could identify regulatory molecules needed to overcome resistance and enhance efficacy in CAR T-cell therapies.
In this review, we look at the status of immune ACT, discussing some of the challenges and solutions that the current therapies are facing. We will describe how single-cell technologies and analysis of large-scale data could provide some answers to those issues and how Mogrify®, using its proprietary direct cell conversion technology, is able to tackle some of the issues associated with immune ACT.
Translation insight
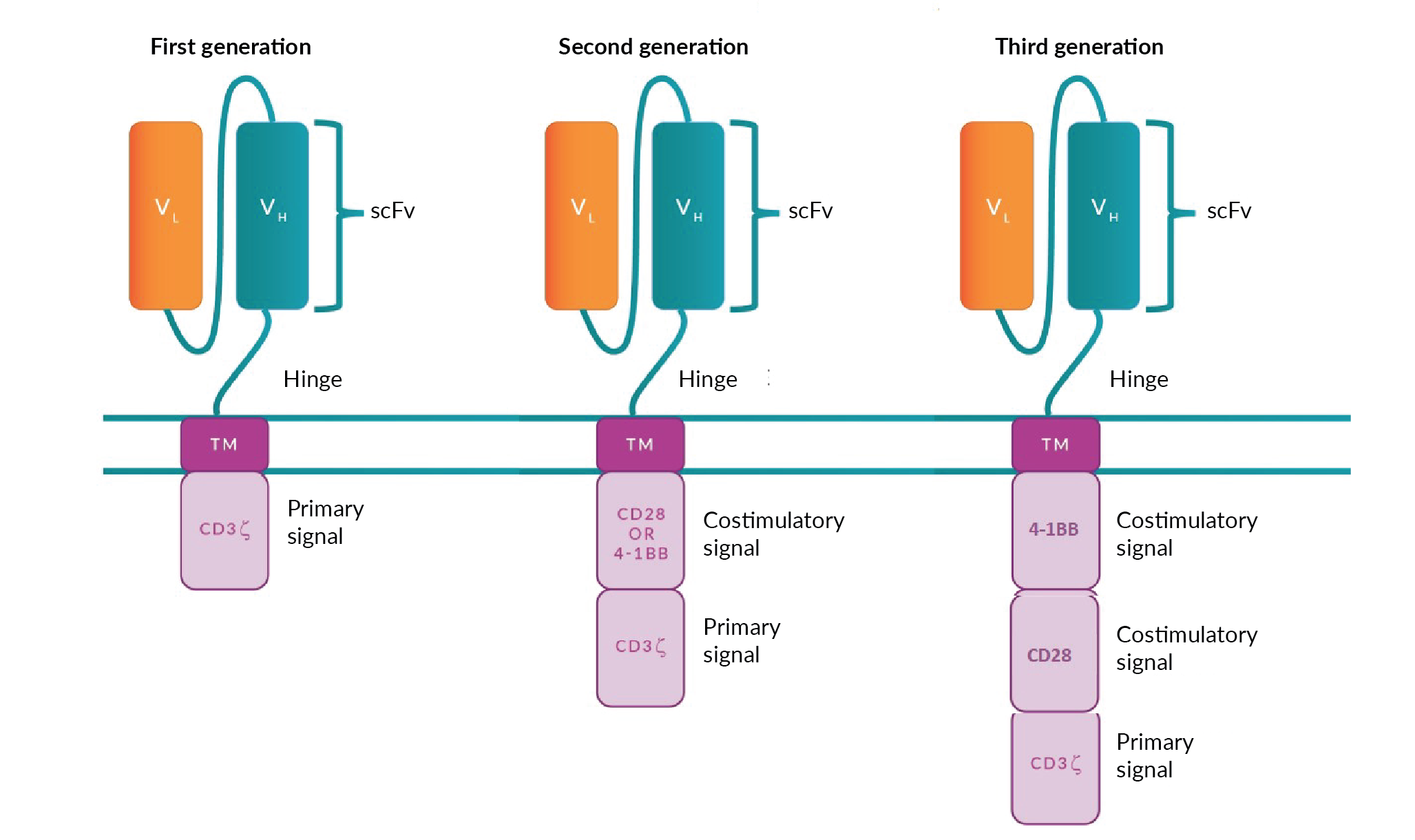
Three cell therapy products based on CAR T cells have so far reached the immuno-oncology market: YESCARTA® (axicabtagene ciloleucel) and TECARTUS™ (brexucabtagene autoleucel) from Kite Pharma and Gilead Sciences; and KYMRIAH® (tisagenlecleucel) from Novartis. KYMRIAH®, TECARTUS™ and YESCARTA® target CD19 which is expressed on malignant, as well as normal, B cells in h[ALL). Generally employed after two or more lines of systemic therapy], [these therapies have elicited complete and lasting tumor regression in up to 40% of patients][4]Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48., [5]Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54., [12]Wang M, Munoz J, Goy A et al. KTE-X19, an Anti-CD19 Chimeric Antigen Receptor (CAR) T Cell Therapy, in Patients (Pts) with Relapsed/Refractory Mantle Cell Lymphoma (R/R MCL): Results of the Phase 2 ZUMA-2 Study. Biol. Blood Marrow Transplant. 2020; 26: S1.. These are autologous therapies, meaning that the patient is both the donor and the recipient of the product. Although this approach has the advantage of avoiding GvHD, there are safety considerations related to unwanted toxicities that may develop following CAR T-cell infusion, such as cytokine release syndrome (CRS), neurological toxicities, ‘on-target/off-tumor’ recognition, and anaphylaxis. CRS occurs in most patients receiving CAR T-cell therapy and based on accumulated experience through many clinical trials, clinical risk management protocols have been put in place so that toxicity is graded according to clinical symptoms and managed pharmacologically [13]Riegler LL, Jones GP, Lee DW. Current approaches in the grading and management of cytokine release syndrome after chimeric antigen receptor T-cell therapy. Ther. Clin, Risk Manag. 2019; 15: 323–35., [14]Bonifant CL, Jackson HJ, Brentjens RJ, Curran KJ. Toxicity and management in CAR T-cell therapy. Mol. Ther. - Oncolytics 2016; 3: 16011.. Scientists are already working towards a new solution to this problem such as the introduction of ‘suicide genes’ into the CAR construct, so that CAR T cells can be selectively depleted [15]Yu S, Yi M, Qin S, Wu K. Next generation chimeric antigen receptor T cells: Safety strategies to overcome toxicity. Mol. Cancer 2019; 18. or turned off [16]Giordano-Attianese G, Gainza P, Gray-Gaillard E et al. A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy. Nat. Biotechnol. 2020; 38: 426–38. if neurotoxicity and CRS are observed in the patient. The use of safety mechanisms may become particularly relevant for allogeneic therapies, where the donor and the recipient (the patient) are two different individuals. In these conditions, GvHD is an unwanted complication likely to occur depending on the degree of Human Antigen Leukocyte (HLA)-mismatch between the donor and the recipient [17]Kanda J. Effect of HLA mismatch on acute graft-versus-host disease. Int. J. Hematol. 2013; 98: 300–8..
The success of KYMRIAH®, TECARTUS™ and YESCARTA® CD19 CAR T-cell therapies has led to exceptional growth in the number of other CAR T-cell therapies targeting this same antigen. Figure 3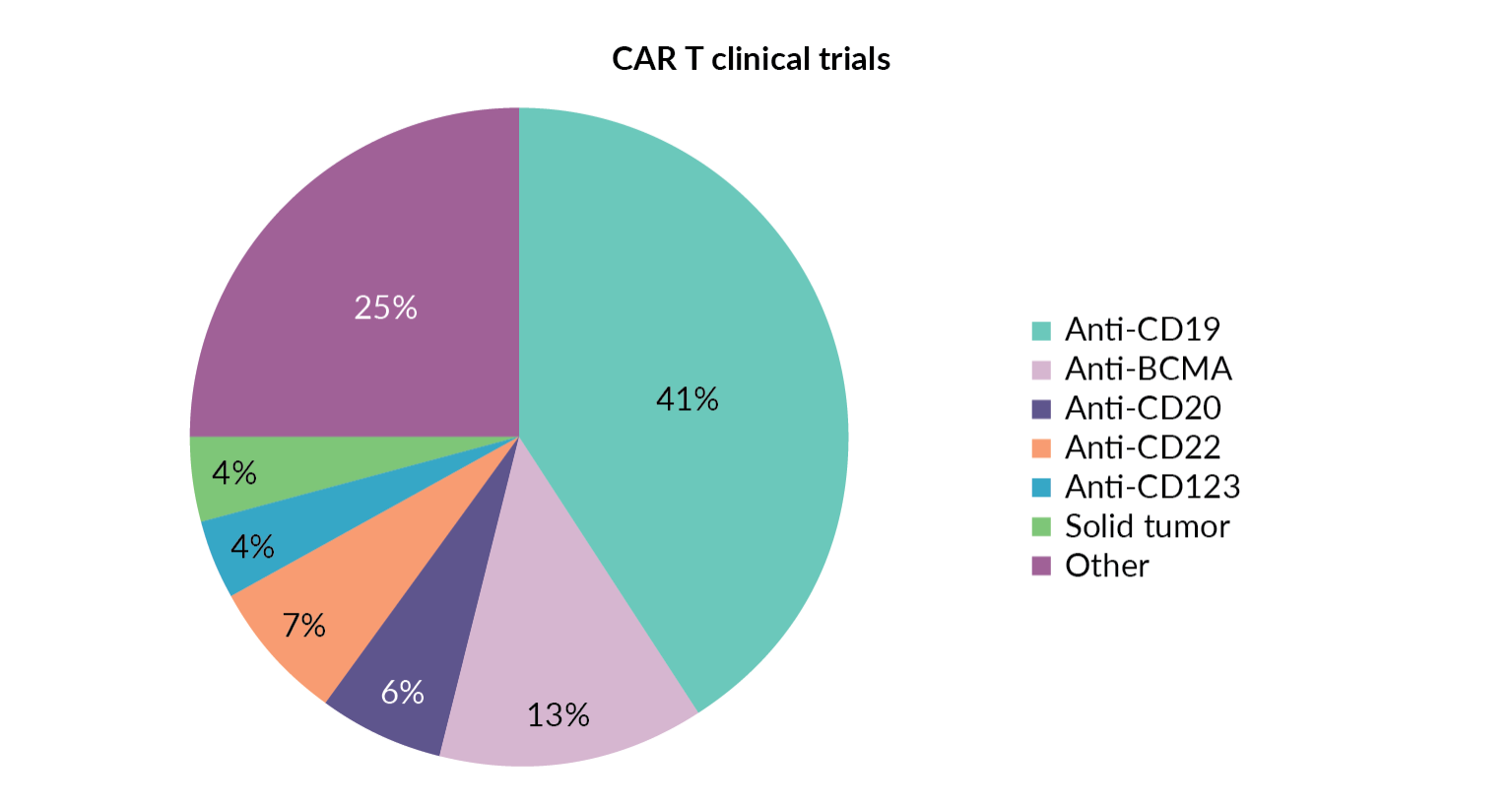
Solid tumors represent a highly challenging environment, as they involve many different cell types that promote, sustain, and protect the growth of the tumor mass via several mechanisms. Myeloid-derived suppressor cells, tumor-associated macrophages, and regulatory T cells generate a suppressive microenvironment by releasing cytokines like IL-10 and upregulating surface markers that inhibit T cell activation. The cells that manage to infiltrate the tumor microenvironment are put on idle by these coercive actions, but often T cells are simply spatially excluded from the tumor. Several mechanisms acting at once may have to be put in place to subvert the tumor microenvironment. Many preclinical studies have shown that CAR T cells genetically modified to secret cytokines (e.g. IL-12, IL-15 and IL-8) could enhance T cell proliferation and anti-tumor activity [18]Pegram HJ, Lee JC, Hayman EG et al. Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood 2012; 119: 4133–41., [19]Hu B, Ren J, Luo Y et al. Augmentation of Antitumor Immunity by Human and Mouse CAR T Cells Secreting IL-18. Cell Rep. 2017; 20: 3025–33.. Moreover, the genetic insertion of chemokine receptors into CAR T cells could lead to an increase of T cell infiltration into the tumor. It has been shown that the expression of CCR2 in CAR T cells increases tumor infiltration and anti-tumor efficacy in preclinical models [20]Moon EK, Carpenito C, Sun J et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin. Cancer Res. 2011; 17: 4719–30.Moon EK, Carpenito C, Sun J et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin. Cancer Res. 2011; 17: 4719–30.. Therefore, multiple mechanisms can be generated to allow the infiltration of an armored CAR T cell into a solid tumor and abrogate the suppressive tumor microenvironment. An example would be to have small molecules that induce endogenous CCR2 expression as an alternative for the transduction of CCR2 gene in mesothelin CAR T cells [20]Moon EK, Carpenito C, Sun J et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin. Cancer Res. 2011; 17: 4719–30.Moon EK, Carpenito C, Sun J et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin. Cancer Res. 2011; 17: 4719–30.. This could increase the safety of CAR T-cell therapy by eliminating any risks related to the insertion of the CCR2 gene. While this technology would possibly be increasing the efficacy of the products, as a result of the boosted T cells’ activity, it will likely increase the likelihood of remission for patients on their last line of treatment (CAR T cell therapy only being prescribed following several rounds of chemotherapy and monoclonal antibody immunotherapies) by using it in combination with checkpoint inhibitors (reviewed in Titov et al. [21]Titov A, Valiullina A, Zmievskaya E et al. Advancing CAR T-cell therapy for solid tumors: Lessons learned from lymphoma treatment. Cancers (Basel) 2020; 12: 1–22.).
On the other hand, the infiltration of a solid tumor may require the use of CAR cell therapies based on alternative cell types to conventional αβT cells. Interest is growing in exploring the potential of other immune cells, including natural killer (NK) cells [22]Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53.Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53.Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53., gamma delta (γδ) T cells [23]Rozenbaum M, Meir A, Aharony Y, Itzhaki O, Schachter J. Gamma-Delta CAR-T Cells Show CAR-Directed and Independent Activity Against Leukemia. Front. Immunol. 2020; 11: 1–8.Rozenbaum M, Meir A, Aharony Y, Itzhaki O, Schachter J. Gamma-Delta CAR-T Cells Show CAR-Directed and Independent Activity Against Leukemia. Front. Immunol. 2020; 11: 1–8. and macrophages [24]Klichinsky M, Ruella M, Shestova O et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020; 1–7.Klichinsky M, Ruella M, Shestova O et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020; 1–7.. All of these cell types have been armed with CARs to test their efficacy in treating solid tumors [22]Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53.Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53.Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53., [23]Rozenbaum M, Meir A, Aharony Y, Itzhaki O, Schachter J. Gamma-Delta CAR-T Cells Show CAR-Directed and Independent Activity Against Leukemia. Front. Immunol. 2020; 11: 1–8.Rozenbaum M, Meir A, Aharony Y, Itzhaki O, Schachter J. Gamma-Delta CAR-T Cells Show CAR-Directed and Independent Activity Against Leukemia. Front. Immunol. 2020; 11: 1–8., [24]Klichinsky M, Ruella M, Shestova O et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020; 1–7.Klichinsky M, Ruella M, Shestova O et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020; 1–7.. NK cells are particularly suitable for immune ACT as they can acquire antigen specificity via the CAR technology, while still retaining their natural cytotoxicity through their ability to recognize target cells via the detection of lack of HLA expression – referred to as ‘missing self-recognition’. Recently, HLA-mismatched anti-CD19 CAR NK cells derived from cord blood were administered to 11 patients with relapsed or refractory CD19-positive lymphoid tumors. It was observed that 7 of the patients had complete remission and without any side effects (cytokine release syndrome, neurotoxicity, or GvHD) [22]Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53.Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53.Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53.. For all of the reasons listed above, NK cells are currently viewed as a good candidate cell type for allogeneic therapies. However, certain challenges remain when working with NK cells for immune ACT. For example, it is difficult to scale up NK manufacturing to support ‘off-the-shelf’ allogeneic treatments [25]Fang F, Xiao W, Tian Z. Challenges of NK cell-based immunotherapy in the new era. Front. Med. 2018; 12: 440–50.. In vitro expansion is necessary for any NK cell-based cell therapy as the cells constitute only 5–15% of peripheral blood monocyte cells (PBMCs). Alternatively, NK cells can be differentiated from cord blood or stem cells. This could reduce variability between batches and increase the quality of the cell product, but in this case NK cells must be differentiated as well as expanded. Protocols for expansion of NK cells rely on cytokines and/or feeder cells, but overall in vitro expansion of NK cells tends to be modest, and often NK cells require an additional in vivo maturation step to acquire full functionality. These complexities have pushed researchers to search for alternative sources, such as the NK-92 cell line, which has been used in clinical trials, and was most recently engineered to express CARs directed towards liquid and solid tumors [26]Tonn T, Schwabe D, Klingemann HG et al. Treatment of patients with advanced cancer with the natural killer cell line NK-92. Cytotherapy 2013; 15: 1563–70., [27]Williams BA, Law AD, Routy B et al. A phase I trial of NK-92 cells for refractory hematological malignancies relapsing after autologous hematopoietic cell transplantation shows safety and evidence of efficacy. Oncotarget 2017; 8: 89256–68.. Although safety and pre-clinical data for efficacy [28]Zhang C, Burger MC, Jennewein L et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl Cancer Inst. 2016; 108: 1–12. have been demonstrated, clinical efficacy is still to be confirmed, and the cells have to be irradiated before use, greatly shortening their lifespan after transplant.
An alternative approach to CARs is the generation of T cells in which the α and β chains of a T cell receptor (TCR) specific for a cancer antigen or neoantigen are expressed as an addition to, or in replacement of, the endogenous TCR [29]He Q, Jiang X, Zhou X, Weng J. Targeting cancers through TCR-peptide/MHC interactions. J. Hematol. Oncol. 2019; 12: 1–17.. Such α and β chains are usually identified from T cell clones enriched in patient biopsies. T cells genetically engineered to express the chosen TCR recognize the cancer antigens through the classical antigen presentation pathway, which processes cytoplasmic as well as surface proteins. To be effective, TCR engineered T cells rely on antigen presentation in the tumor microenvironment, which is often downregulated in cancer cells as an escape mechanism. HLA matching between the donor and the recipient is also required, making this an autologous ACT, although some degree of HLA-mismatching may be tolerated and could potentially be used in allogeneic ACT. The number of clinical trials that employ TCR-engineered T cells in cancer treatment is much lower compared to those for CAR T cells [30]Ping Y, Liu C, Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell 2018; 9: 254–66., perhaps due to the complexity of identifying good ‘universal’ target antigens, and the challenges to produce a TCR with the optimal affinity and avidity for the cognate antigen.
The field of cell therapy for immuno-oncology is rich in possibilities for both autologous and allogeneic treatments, using different modalities such as CARs or TCR engineered receptors, and different cell types, like T cells, NK cells, and others [8]Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.. Moreover, the synergy of combinatory approaches such as cell therapy and immune checkpoint inhibitors may enable the immune system to disrupt and destroy the tumor microenvironment. Much knowledge will be obtained from the data generated by ongoing and new clinical trials, providing a solid basis for ever safer and more effective cell-based therapies. Moreover, the lessons learned in immuno-oncology are now rapidly applied to GvHD and to the other side of the coin in immunology, autoimmunity. The exciting possibility to use technologies like CARs to induce immunological tolerance to treat and prevent organ rejection, or to restore immunological balance in autoimmune diseases, has already started to be explored, and promises to have a big impact on the lives of many people living with chronic conditions.
Manufacturing aspects of cell therapy
Traditionally, pharmaceutical development follows a defined pathway that covers all stages of a therapeutic product. This is summarized by the first phase of discovery and development, followed by preclinical and clinical research phases that lead to drug approval, then by post-market safety monitoring. Starting from the clinical research phase, all processes must adhere to Good Manufacturing Practice (GMP), which ensures that fully characterized, controlled and consistent manufacturing processes are in place to guarantee the safety and efficacy of products in accordance with pre-determined quality standards.
ACT has questioned and even bypassed traditional pharmaceutical development, challenging the status quo. In cell therapy, early development has mainly taken place in the clinic, frequently under hospital exemption. Moreover, the preclinical phase is challenging due to the lack of relevant animal models that truly recapitulate human disease. Finally, the first therapies to reach the immuno-oncology market are autologous, made from cells collected from one patient for the treatment of the same patient, epitomizing the concept of ‘personalized medicine’ [4]Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48.Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48., [5]Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54.. This model does not fit with production scale-up processes in the same way as, for example, small molecules or even biologics. Among the many variables that characterize this model, the quantity and quality of the starting material are notable, as these heavily depend on each patient’s medical history. ACT in immuno-oncology is currently approved for patients who have undergone two or more lines of systemic therapy, meaning that the starting material from which the therapeutic product is manufactured can be extremely variable between patients, and even within the same patient if the cell therapy product has to be made on more than one occasion for additional dosing. Depending on the quality and quantity of the starting material, the manufacturing process must be adjusted for each batch.
In addition, the manufacturing process is often still manual, although appropriate automated solutions have been developed and have started to be implemented. In the current ACT setting, the clinical and manufacturing teams have to work closely to be able to synchronize product manufacturing and patient treatment (reviewed in [31]Levine BL, Miskin J, Wonnacott K, Keir C. Global Manufacturing of CAR T Cell Therapy. Mol. Ther. – Methods Clin. Dev. 2017; 4: 92–101.Levine BL, Miskin J, Wonnacott K, Keir C. Global Manufacturing of CAR T Cell Therapy. Mol. Ther. – Methods Clin. Dev. 2017; 4: 92–101.). While the manufacturing team generates the cell product, the clinical team has to assist the patient to undertake a conditioning treatment to ‘make space’ for the ACT after infusion. To accommodate this model, ACT manufacturing has been kept in close proximity to the clinic, often with manufacturing suites located at the hospital site or nearby. Analytical, quality control and quality assurance teams are also likely to be located close to the hospital for the same reasons. This modus operandi is typically more complicated and more expensive to manage administratively, and it is referred to as ‘scaling out’. Scaling out requires a different manufacturing set up compared to classical drug manufacturing, and this model has been adopted by most cell therapy companies. Bigger companies have started to move away from scaling out and adopted the classical ‘scaling up’ model by setting up manufacturing centers in strategic locations served by appropriated transport facilities. In this case, logistics and operations become even more crucial to the completion of the tight vein-to-vein turnaround allocated for manufacturing and product release. As this sometimes involves shipping items over considerable distances, suitable transportation systems have been developed to guarantee that cell therapy products are delivered safely and on time [31]Levine BL, Miskin J, Wonnacott K, Keir C. Global Manufacturing of CAR T Cell Therapy. Mol. Ther. – Methods Clin. Dev. 2017; 4: 92–101.Levine BL, Miskin J, Wonnacott K, Keir C. Global Manufacturing of CAR T Cell Therapy. Mol. Ther. – Methods Clin. Dev. 2017; 4: 92–101.. This also demands that the chain of custody and identity of the product are maintained throughout the entire process, as a failure to document the identity and integrity of the product could have fatal consequences for the patient.
Currently, one of the main drawbacks of ACT is its high cost, which is reported to be $475,000/dose for YESCARTA®. Due to the ‘ad hoc’ manual or semi-automated manufacturing process, one of the major contributors to the cost of goods is labor [32]Spink K, Steinsapir A. The long road to affordability: a cost of goods analysis for an autologous CAR-T process. Cell Gene Ther. Insights 2018; 4: 1105–16.. The rate of optimization, and implementation of automation and process simplification will therefore determine the speed at which ACT becomes affordable. Significant advances have already been made with the development of modular automated systems that reduce the ‘hands-on’ time required for product manufacturing. Nevertheless, due to the limitation imposed by the fact that one batch is made for one patient, it is unlikely that the cost of cell therapy will suddenly drop. The cost of GMP-grade raw materials is also high, but it is likely that as the industry continues to grow new solutions will become available. An example is illustrated by the shortage of animal-derived serum for the growing cell therapy sector that was forecasted in 2012 [33]Brindley DA, Davie NL, Culme-Seymour EJ, Mason C, Smith DW, Rowley JA. Peak serum: Implications of serum supply for cell therapy manufacturing. Regen. Med. 2012; 7: 7–13. – eight years later, although serum is still in high demand, serum-free alternatives are available and have already been implemented by some.
Most of the issues described above relate to autologous cell therapy, mainly due to the personalized nature of the treatment. In contrast, the manufacture of allogeneic products is less challenging [8]Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.. The paradigm of ‘one batch for one patient’ that characterizes autologous therapy is replaced with ‘one batch for several patients’ in allogeneic therapies. In allogeneic therapy, the pathway for manufacturing is more aligned with the traditional model of scaling up, mainly thanks to the uncoupling of the vein-to vein turnaround. To meet demands, innovative and more efficient systems are being developed, such as the use of suspension cells instead of adherent cells for viral production, or large volume bioincubators and automated closed systems. One of the major challenges for scaling up off-the-shelf manufacturing is the availability of the large volume and consistently high quality of cells required. Innovative solutions and beginning to be identified and some have now been developed and started to be tested in clinical trials.
The discovery of in vitro methods for differentiating as well as modifying the differentiation status of cells has come to the rescue of cell therapy manufacturing. A good source of cells for further differentiation and genetic manipulation are embryonic stem cells (ESCs), for which substantial knowledge has already been accumulated in other fields, mesenchymal stem/stromal cells (MSCs) and induced pluripotent stem cells (iPSCs). Figure 4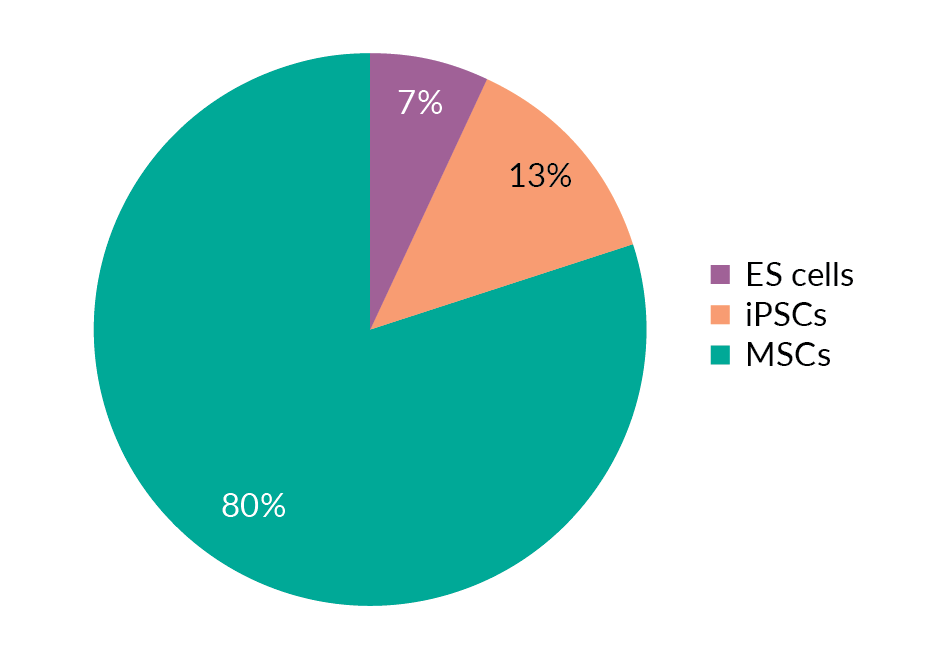
Although these cell types are currently used mainly in transplantation and regenerative medicine, their usage as starting materials to manufacture clinical-grade cell therapy products is also foreseeable in the near future, which will require the development of cell differentiation methods compliant with GMP. The availability of iPSCs has also opened new possibilities in ACT for immuno-oncology. The employment of iPSCs as starting material to be differentiated into the desired cell type will allow the generation of large batches of identical cells obtained from suitable donors and enable the manufacturing of ‘off-the-shelf’ cell therapies. Moreover, research around the world is concentrating on developing even more advanced tools, such as a hypoimmunogenic universal donor cell line [34]Lanza R, Russell DW, Nagy A. Engineering universal cells that evade immune detection. Nat. Rev. Immunol. 2019; 19: 723–33.. The latter is as challenging as it is desirable, and could provide a ‘blank canvas’ on which to add further properties to create a new ‘artificial cell’ that does not cause GvHD, and is poised to recognize and kill tumors with high specificity as well as safety.
The emerging landscape of next-generation modalities in single-cell analysis & their application in immune cellular therapies
Many methods have been used to characterize and understand immune cells over the past century, from morphology and tissue distribution through to modern flow cytometry capable of measuring the expression of >20 proteins. This has given us a deep understanding of the markers and functions of different cell types in a range of tissues and in response to different stimuli. This synergy between immunology and emerging technologies ensures that our paradigms are continually updated to consider new methods and information.
Biology has often been limited by the fact that approaches that give information about many genes or proteins are limited to a few samples of large numbers of cells. However, recent years have seen an explosion of technologies available to study cells at the single-cell level, and the application of these technologies to immune cells [35]Efremova M, Vento-Tormo R, Park J-E, Teichmann SA, James KR. Immunology in the Era of Single-Cell Technologies. Annu. Rev. Immunol. 2020; 38. , [36]See P, Dutertre CA, Chen J et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science (80) 2017; 356. See P, Dutertre CA, Chen J et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science (80) 2017; 356. , [37]Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science (80). 2017; 358: 58–63., [38]Yost KE, Chang HY, Satpathy AT. Tracking the immune response with single-cell genomics. Vaccine 2019; 38(28): 4487–90.Yost KE, Chang HY, Satpathy AT. Tracking the immune response with single-cell genomics. Vaccine 2019; 38(28): 4487–90., [39]Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018; 18: 35–45.. These have largely been based around sequencing technologies, including transcriptome profiling using RNA sequencing and single-cell RNAseq (scRNAseq) [40]Method of the Year 2013. Nat. Methods. 2014; 11: 1., and epigenomic studies using assay for transposase-accessible chromatin (ATAC)-seq [41]Lareau CA, Duarte FM, Chew JG et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 2019; 37: 916–24., [42]Satpathy AT, Saligrama N, Buenrostro JD et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 2018; 24: 580–90.Satpathy AT, Saligrama N, Buenrostro JD et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 2018; 24: 580–90., [43]Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015; 2015: 21.29.1–21.29.9., [44]Cusanovich DA, Daza R, Adey A et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science (80) 2015; 348: 910–4.. Additionally, cytometry by time-of-flight (CyTOF) [45]Bendall SC, Simonds EF, Qiu P et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science (80) 2011; 332: 687–96. has expanded the number of proteins we can analyze compared with flow cytometry by tagging antibodies with heavy metals and passing stained cells through a mass spectrometer to detect protein expression by molecular weight, rather than fluorescence. These methods are now frequently used in combination on the same cells [46]Packer J, Trapnell C. Single-Cell Multi-omics: An Engine for New Quantitative Models of Gene Regulation. Trends Genet. 2018; 34: 653–65., [47]Cao J, Cusanovich DA, Ramani V et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science (80) 2018; 361: 1380–5., for example, to look at changes in the transcriptome [48]Parekh U, Wu Y, Zhao D et al. Mapping Cellular Reprogramming via Pooled Overexpression Screens with Paired Fitness and Single-Cell RNA-Sequencing Readout. Cell Syst. 2018; 7: 548–555.e8.Parekh U, Wu Y, Zhao D et al. Mapping Cellular Reprogramming via Pooled Overexpression Screens with Paired Fitness and Single-Cell RNA-Sequencing Readout. Cell Syst. 2018; 7: 548–555.e8., [49]Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177.Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177.Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177. or chromatin accessibility [50]Rubin AJ, Parker KR, Satpathy AT et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 2019; 176: 361–376.e17. in response to gene knockdown in CRISPR screens, or coupling protein expression with transcriptomics by using DNA-tagged antibodies for immunophenotyping (CITEseq) [51]Stoeckius M, Hafemeister C, Stephenson W et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017; 14: 865–8.. There are also many resources comparing these different technologies and methods to analyze data [52]Mereu E, Lafzi A, Moutinho C et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 2020; 1–9., [53]Stuart T, Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019; 20: 257–72..
These technologies have allowed researchers to assimilate a lot of new knowledge without a priori assumptions, such as screening for immune cell subsets present in a variety of different tissues and species [54]Paul F, Arkin Y, Giladi A et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 2015; 163: 1663–77.Paul F, Arkin Y, Giladi A et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 2015; 163: 1663–77. and how that changes in autoimmune diseases [55]Dutertre CA, Becht E, Irac SE et al. Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity 2019; 51: 573–589.e8., harmonizing the markers used to identify cell types across different species [56]Guilliams M, Dutertre CA, Scott CL et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity 2016; 45: 669–84., and to better understand the ontogeny of immune subsets [36]See P, Dutertre CA, Chen J et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science (80) 2017; 356. See P, Dutertre CA, Chen J et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science (80) 2017; 356. , [54]Paul F, Arkin Y, Giladi A et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 2015; 163: 1663–77.Paul F, Arkin Y, Giladi A et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 2015; 163: 1663–77.. There are efforts underway to map the entire human body at single-cell resolution using these technologies, in the Human Cell Atlas project [57]Regev A, Teichmann SA, Lander ES et al. The human cell atlas. Elife 2017; 6. .
Due to the somatic recombination that the T and B cell receptor genes undergo during differentiation, we can also track cell clones through TCR and BCR sequencing at the single-cell level (so-called V(D)J sequencing) [58]Stubbington MJT, Lönnberg T, Proserpio V et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 2016; 13: 329–32., [59]Lindeman I, Emerton G, Mamanova L et al. BraCeR: B-cell-receptor reconstruction and clonality inference from single-cell RNA-seq. Nat. Methods 2018; 15: 563–5.. This allows the monitoring of clonal evolution in infection and disease settings, and through aging processes. MHC-dextramers tagged with DNA barcodes have also been used to probe the antigen-specificity of T cells [60]Setliff I, Shiakolas AR, Pilewski KA et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell 2019; 179: 1636–1646.e15.. Coupling transcriptomics with V(D)J sequencing, CITEseq, CRISPR screening and antigen-specificity has allowed up to five modalities to be analyzed from the same cells using next-generation sequencing [61]Mimitou EP, Cheng A, Montalbano A et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 2019; 16: 409–12.. These technologies are allowing us to interrogate the complexities of the immune system at unprecedented scale and resolution [38]Yost KE, Chang HY, Satpathy AT. Tracking the immune response with single-cell genomics. Vaccine 2019; 38(28): 4487–90.Yost KE, Chang HY, Satpathy AT. Tracking the immune response with single-cell genomics. Vaccine 2019; 38(28): 4487–90..
Selectively programing cell fate
A key aim of these new technologies is to better understand cell ontogeny and mechanisms of cell fate decisions [62]Watcham S, Kucinski I, Gottgens B. New insights into hematopoietic differentiation landscapes from single-cell RNA sequencing. Blood 2019; 133: 1415–26., [63]Pijuan-Sala B, Guibentif C, Göttgens B. Single-cell transcriptional profiling: A window into embryonic cell-type specification. Nat. Rev. Mol. Cell Biol. 2018; 19: 399–412., [64]Trapnell C. Defining cell types and states with single-cell genomics. Genome Res. 2015; 25: 1491–8., accompanied by computational techniques for identifying differentiation trajectories [65]La Manno G, Soldatov R, Zeisel A et al. RNA velocity of single cells. Nature 2018; 560: 494–8., [66]Trapnell C, Cacchiarelli D, Grimsby J et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32: 381–6., [67]Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 2016; 13: 845–8.. This could translate into improved cell therapies by directed differentiation from pluripotent cells by identifying key transcription factors (TFs) or signaling pathways required during differentiation. This is of particular interest in immuno-oncology where autologous CAR T and NK cell products are expensive and time-consuming to make, and fraught with issues, so the search for allogeneic ‘off-the-shelf’ alternatives is intensive with many companies developing iPSC-derived products [8]Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99.Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99., [68]Li Y, Hermanson DL, Moriarity BS, Kaufman DS. Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell Stem Cell 2018; 23: 181-192.e5.. Many of these protocols are also very long and costly as they replicate normal human development, so there is interest in bypassing normal differentiation by overexpressing TFs, which are often viewed as the master regulators of cell fate. Additionally, cells derived from ESCs or iPSCs often have an immature, fetal phenotype and lack full adult function, so improvements are required to fully realize their potential. Papers describing the reprograming of cell fate using the overexpression of transcription factors have spanned decades [69]Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987; 51: 987–1000., [70]Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006; 126: 663–76., [71]Rosa FF, Pires CF, Kurochkin I et al. Direct reprogramming of fibroblasts into antigen-presenting dendritic cells. Sci. Immunol. 2018; 3: 1–16., but it has been challenging to identify optimal combinations of TFs for conversions, usually involving trial and error. Rational, data-driven selection of TFs and scalable screening methods are required to accelerate discovery in this area, and new technologies are aiding this process.
One strategy, Reprogram-Seq [72]Duan J, Li B, Bhakta M et al. Rational Reprogramming of Cellular States by Combinatorial Perturbation. Cell Rep. 2019; 27: 3486-3499.e6., predicts candidate TFs by identifying genes that are differentially expressed between source and target cells in scRNAseq data. Pools of TFs are then overexpressed in the source cells, such that each cell will receive a different, random combination of factors. The converted cells are then surveyed using scRNAseq for transcriptional signatures that match the target cell of interest, and the transgenes that were overexpressed can be identified as they lack a 3' UTR compared with the endogenous transcripts for the same factor. MOGRIFY® can efficiently navigate a combinatorial space of >500 billion possible TF combinations to find the optimal set of TFs controlling the genetic programs required to be switched for a given cell conversion [9]Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.. This results in few transcription factors with the highest and non-overlapping regulatory influence. Initially, the algorithm was run to generate predictions for cell conversions and has successfully demonstrated multiple cell conversions, including keratinocytes from fibroblasts, and endothelial cells from keratinocytes [9]Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5..
An alternative method, SEUSS (scalable functional screening by sequencing), addressed the effect of overexpressing a library of 61 developmentally important TFs in iPSCs, including modified versions of genes that could not be predicted by Reprogram-seq [48]Parekh U, Wu Y, Zhao D et al. Mapping Cellular Reprogramming via Pooled Overexpression Screens with Paired Fitness and Single-Cell RNA-Sequencing Readout. Cell Syst. 2018; 7: 548–555.e8.Parekh U, Wu Y, Zhao D et al. Mapping Cellular Reprogramming via Pooled Overexpression Screens with Paired Fitness and Single-Cell RNA-Sequencing Readout. Cell Syst. 2018; 7: 548–555.e8.. The effect of TF overexpression was screened by scRNAseq, coupled with a fitness readout of cell growth in multiple culture conditions. In contrast to Reprogram-seq, a barcode associated with each transgene was used to identify useful TFs from the sequencing data. These data were used to construct a genetic co-regulatory network based on transcriptomic changes, identifying key factors for early fate specification. This included identifying ETV2 as a reprograming factor for an endothelial-like state, a useful validation of the method as this TF was already known as an important regulator of early blood and endothelial specification during embryogenesis.
While these are important proof of concept studies, it is difficult to screen the whole transcriptome this way as the number of possible TF combinations scales rapidly, and it would be prohibitively expensive to sequence and analyze the required number of cells. It is also important to consider that each converted cell in a pooled screen is not an independent experiment, and the effects of paracrine and juxtracrine signaling may be significant. Better computational methods to narrow down the set of TFs used will have great value. CellNet assesses the quality of cell differentiation or conversion experiments by comparing transcriptome information to reference data and identifying genes that can be modulated to enhance conversion [73]Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902.Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902.Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902.. By contrast, MOGRIFY® can predict de novo the combination of TFs required to induce direct cell conversion from any cell type to any other cell type using a combination of transcriptome data, protein-protein, and protein-DNA interaction databases to identify the network of genes to activate, and the TFs that regulate them, removing the guesswork from direct cell conversions [9]Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.. Most screens also focus on overexpression of TFs as this gives strong and stable expression of the gene of interest and control over the isoforms used. However, the regulatory impact of non-coding RNAs and the need to downregulate particular genes are important factors that should be included in future studies. An overview of the different algorithms and their approach to identifying TFs for cell conversion is summarized in Table 1 and in [9]Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5., [73]Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902.Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902.Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902., [74]Kamaraj US, Gough J, Polo JM, Petretto E, Rackham OJL. Computational methods for direct cell conversion. Cell Cycle 2016; 15: 3343–54., [75]D’Alessio AC, Fan ZP, Wert KJ et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015; 5: 763–D’Alessio AC, Fan ZP, Wert KJ et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015; 5: 763–, [76]Okawa S, Saltó C, Ravichandran S et al. Transcriptional synergy as an emergent property defining cell subpopulation identity enables population shift. Nat. Commun. 2018; 9: 1–10.Okawa S, Saltó C, Ravichandran S et al. Transcriptional synergy as an emergent property defining cell subpopulation identity enables population shift. Nat. Commun. 2018; 9: 1–10..
| Table 1. Summary of methods. | ||||||
| Platform name | Computational steps for TF prediction | Reference | ||||
| Input data type required | Data used for identifying cell identity profiles | Strategy used to identify TF influence | Criteria to prioritize TFs | Prediction | ||
| MOGRIFY® | RNA-Seq, CAGE | Fantom CAGE dataset (274 cell types) | Build cell type-specific regulatory network of TF and target genes | Cell type specificity in the target cell of the regulatory network and upstream TF regulators | Non-redundant set of core TFs | Rackham et al.[9]Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5. |
| JSD | Microarray | GEO Microarray database (233 cell types) | Target cell versus background cell types | By JSD specific score | Core of 10 TFs | D’Alessio et al.[75]D’Alessio AC, Fan ZP, Wert KJ et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015; 5: 763–D’Alessio AC, Fan ZP, Wert KJ et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015; 5: 763– |
| CellNet | Microarray | GEO Microarray database (16 cell types) | Differential expression | By number of regulated gene and TF expression fold changes | Target Cell specific network | Morris et al.[73]Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902.Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902.Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902. |
| TranSyn | Single-cell RNA-Seq | Single cell RNA-Seq data clustered by subpopulation | Multivariate mutual information (MMI) starting from the most expressed TFs | Maximize MMI value | List of TFs | Okawa et al.[76]Okawa S, Saltó C, Ravichandran S et al. Transcriptional synergy as an emergent property defining cell subpopulation identity enables population shift. Nat. Commun. 2018; 9: 1–10.Okawa S, Saltó C, Ravichandran S et al. Transcriptional synergy as an emergent property defining cell subpopulation identity enables population shift. Nat. Commun. 2018; 9: 1–10. |
| Each row represents the method for predicting TFs in transdifferentiation. Each column represents the computational stages involved in the TFs set prediction which are input requirement, generation of differential expression profile, identifying the influence of each TF in cell conversion, criteria to prioritize the TFs and finally the predictions. | ||||||
New methodologies also offer the potential to better understand the role of the tissue microenvironment in development, which could aid in improving culture conditions for directed differentiation or direct cell conversions. A paper from the Human Cell Atlas project surveyed the dynamics of human thymic development from the fetus through to >30 years postnatal life [77]Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.. T cell development requires a complex interaction of T cells with the thymic stroma and dendritic cells to direct fate towards the multiple functional lineages, in combination with the rearrangement of the TCR genes that define antigen specificity. The study included scRNAseq analysis of T cells and other immune and stromal cells, to show how such interactions shape T cell development and repertoire, as determined using TCR sequencing. The study observed early emergence of innate-like T lymphocytes (including γδT cells and CD8ɑα+ T cells), with conventional αβT cells developing later [77]Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224., in line with reports that T cells derived in vitro from iPSCs show a tendency towards an innate-like phenotype or do not fully recapitulate the typical properties of their phenotype [78]Themeli M, Rivière I, Sadelain M. New cell sources for T cell engineering and adoptive immunotherapy. Cell Stem Cell 2015; 16: 357–66.. Computational tools were used to predict the trajectory of cell differentiation, identifying waves of TCR recombination and sets of stage-specific TFs regulating differentiation. This analysis is important as recent differentiation protocols have highlighted the importance of TCR expression in differentiation, with iPSCs genetically edited to carry a particular transgenic TCR or CAR undergoing superior differentiation, compared with unedited cells, in the absence of a thymic microenvironment [79]Minagawa A, Yoshikawa T, Yasukawa M et al. Enhancing T Cell Receptor Stability in Rejuvenated iPSC-Derived T Cells Improves Their Use in Cancer Immunotherapy. Cell Stem Cell 2018; 23: 850-858.e4.. To understand the role of stromal and dendritic cells, the study made use of CellPhoneDB, the authors’ previous work that uses a statistical framework and known receptor-ligand pairs to predict enriched cellular interactions from scRNAseq data [80]Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc 2020; 1–23.. This identified chemokine signatures promoting migration of T cells from one area of the thymus to another during differentiation. Single molecule fluorescent in situ hybridization (smFISH), a technique that identifies mRNA expression at single-cell resolution in tissue slices, was used to validate the sub-thymic localization of cell subsets and the predicted intercellular interactions [77]Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224.. Lastly, spatial transcriptomics can allow for cellular transcriptional sequencing in situ, for informing on ACT for solid tumors. While not being strictly at the single-cell level in the case of 10x Visium, when used in conjunction with scRNAseq, the transcriptional signatures can be deconvoluted. These approaches have already been applied to understand the cellular microenvironment of solid tumors [81]Yoosuf N, Navarro JF, Salmén F, Ståhl PL, Daub CO. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res 2020; 22: 1–10., [82]Berglund E, Maaskola J, Schultz N et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018; 9. .
Next-generation sequencing and high throughput data approaches will play a key role in the identification of gene regulators or soluble factors that can increase the quality of ACT products. As an example, MOGRIFY® leverages datasets such as the FANTOM5 consortia data, which uses Cap Analysis of Gene Expression (CAGE) to map the sets of transcripts, transcription factors, promoters and enhancers active in the majority of mammalian primary cell types, making it amenable to in-depth transcriptomic analysis. The technology uses a big-data algorithm to compare gene expression and identify the optimal combination of transcription factors required to directly convert any cell type into any other [9]Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5.. In addition, the same dataset has made it possible to identify enhancers and promoters that are important in T cell and macrophage differentiation by profiling human T cells and monocytes [10]Schmidl C, Hansmann L, Lassmann T et al. The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood 2014; 123: 68–78.Schmidl C, Hansmann L, Lassmann T et al. The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood 2014; 123: 68–78., [11]Schmidl C, Renner K, Peter K et al. Transcription and enhancer profiling in human monocyte subsets. Blood 2014; 123: 90–9.Schmidl C, Renner K, Peter K et al. Transcription and enhancer profiling in human monocyte subsets. Blood 2014; 123: 90–9.. This type of large-scale data could be used to identify the regulatory molecules needed to overcome resistance in ACT, to bypass lengthy differentiation protocols from pluripotent stem cells, and to reduce variability between batches of cellular products. These types of approaches can also be applied to improve infiltration in solid tumors or to identify and engineer switch receptors that transform suppression signals and increase CAR T-cell resistance to the tumor microenvironment.
Multi-omic & screening approaches to tackle T cell exhaustion
As well as interest in programing cell fate, the era of genome engineering also provides opportunities to enhance cell function or overcome roadblocks to cell therapies. A large focus has been placed on circumventing the issue of T cell exhaustion, where repeated stimulation leads to cellular dysfunction and impaired immune response.
In one recent study, T cells from patients with basal cell carcinoma were analyzed by scRNAseq coupled with TCR sequencing before and after treatment with anti-PD-1-antibody. Interestingly, this study identified a spectrum of T cell phenotypes in the tumors, but showed that checkpoint blockade does not reinvigorate tumor-infiltrating leukocytes, as previously thought, but allows novel cell clones to enter the tumor from the circulation [83]Yost KE, Satpathy AT, Wells DK et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019; 25: 1251–9.. Combining TCR sequencing and ATACseq of single cells [42]Satpathy AT, Saligrama N, Buenrostro JD et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 2018; 24: 580–90.Satpathy AT, Saligrama N, Buenrostro JD et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 2018; 24: 580–90. in a similar set of patients identified an enhancer within the PDCD1 locus, encoding PD-1, which becomes activated during exhaustion [84]Satpathy AT, Granja JM, Yost KE et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019; 37: 925–36.. Investigating the TF binding motifs in such regions could help identify targets to regulate exhaustion, in the context of our new understanding of clonal dynamics.
Another study combined a variety of ‘omics techniques to search for factors that could overcome exhaustion [85]Lynn RC, Weber EW, Sotillo E et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019; 576: 293–300.Lynn RC, Weber EW, Sotillo E et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019; 576: 293–300.. T cells expressing different CARs were treated with several different stimuli to induce exhaustion profiles and compared to identify differentially expressed genes driving exhaustion. Exhaustion is associated with epigenetic changes, and scATACseq identified differentially accessible regions of chromatin near exhaustion-associated genes such as CTLA-4 in exhausted T cells, and a decrease in accessibility at genes associated with memory, such as IL7A. DNA motifs for the AP1 complex were enriched among the newly open regions in exhausted T cells. The canonical AP1 complex of c-JUN and c-FOS drives expression of IL2, but can be antagonized by other family members, and such factors were found to be upregulated in the transcriptomes of exhausted cells. Overexpression of c-JUN was shown to restore T cell function, and experiments using modified c-JUN proteins lacking functional domains showed that its interaction with other proteins was important for this function rather than its DNA binding capacity. Additionally, overexpression of c-JUN also rendered the CAR T cells more sensitive to lower levels of antigen, which could help in tumors with low antigen expression and where the selective pressure from CAR T cells leads to antigen down-regulation [85]Lynn RC, Weber EW, Sotillo E et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019; 576: 293–300.Lynn RC, Weber EW, Sotillo E et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019; 576: 293–300.. However, c-JUN is potentially oncogenic, so the potential of such modifications to produce to unwanted side effects in the modified T cells needs to be thoroughly assessed.
These studies illustrate how modern technologies can be used to understand the mechanisms behind phenomena such as T cell exhaustion and inform the rational design of strategies to circumvent these mechanisms. However, while they provide a deeper understanding of exhaustion, they have limited throughput for discovering and validating targets to enhance the therapeutic effect of adoptively transferred cells. The development of CRISPR technology has greatly facilitated genome-wide knockout screens for target discovery across biology. Combining this with scRNAseq has provided a balance between the high-dimensionality of arrayed screens, where knockouts are considered one-by-one, with the throughput of pooled screens [49]Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177.Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177.Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177., [86]Dixit A, Parnas O, Li B et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016; 167: 1853-1866.e17.Dixit A, Parnas O, Li B et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016; 167: 1853-1866.e17.. This technology depends on sequencing either a barcode associated with the CRISPR guide RNA [49]Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177.Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177.Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177., [86]Dixit A, Parnas O, Li B et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016; 167: 1853-1866.e17.Dixit A, Parnas O, Li B et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016; 167: 1853-1866.e17. or by sequencing the guide RNA itself after capture onto microbeads [87]Dong MB, Wang G, Chow RD et al. Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell 2019; 178: 1189–1204.e23.Dong MB, Wang G, Chow RD et al. Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell 2019; 178: 1189–1204.e23.. Several CRISPR knockout screens coupled with scRNAseq and tumor infiltration models have identified regulators of CD8+ T cell fitness in mice, including REGNASE-1 [87]Dong MB, Wang G, Chow RD et al. Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell 2019; 178: 1189–1204.e23.Dong MB, Wang G, Chow RD et al. Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell 2019; 178: 1189–1204.e23., [88]Wei J, Long L, Zheng W et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 2019. , [89]Ye L, Park JJ, Dong MB et al. In vivo CRISPR screening in CD8 T cells with AAV–Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotechnol. 2019; 37: 1302–13..
Roth et al. used a targeted approach to examine both wild type and novel constructs that could be introduced to cells to enhance CD8+ T cell function [90]Roth TL, Li PJ, Blaeschke F et al. Pooled Knockin Targeting for Genome Engineering of Cellular Immunotherapies. Cell 2020; 181: 728–744.e21.. The system used CRISPR-mediated homologous recombination to introduce a transgene carrying a constant transgenic TCR targeting the NY-ESO-1 antigen into the endogenous TRAC locus, along with one of the genes used in the screen, and a transgene-specific barcode. Modified cells were challenged in a number of assays to identify transgenes that could enhance tumor infiltration and cytotoxicity towards NY-ESO-1-expressing cancer cells in humanized mouse models, under stimulation with anti-CD3/CD28, the immunosuppressive cytokine TGFβ, or other factors. After functional readouts, the cells were sampled for scRNAseq and targeted sequencing of the knock-in barcode to couple transgene expression to the transcriptome. Interestingly, while the authors found that knocking in either TCF7 or a synthetic TGFbR2-41BB receptor increased T cell abundance in solid tumors, the latter promoted accumulation of cells expressing key effector cytokines while the TCF7-expressing cells failed to function, highlighting the importance of using multiple functional assays to assess phenotypes. While very interesting, this approach is of course limited to transgenes selected a priori rather than representing an unbiased screening approach.
CRISPR holds great promise, but there are concerns around off-target editing events and long-term safety of edited cells. An important component of clinical studies is, therefore, tracking the fate of adoptively transplanted cells. Results of the first clinical study with CRISPR-edited T cells were published in early 2020 [91]Stadtmauer EA, Fraietta JA, Davis MM et al. CRISPR-engineered T cells in patients with refractory cancer. Science (80- ) 2020; 367: eaba7365.. T cells were modified to remove the endogenous genes encoding the T cell receptor (TRAC and TRBC) and introduce a cancer-specific TCR, a strategy thought to enhance transgene function by preventing the mispairing of endogenous and exogenous TCRα and β chains. The PDCD1 gene was also removed to limit exhaustion and enhance anti-tumor immunity, resulting in a total of three genomic edits. scRNAseq of cells from patient samples was used to assess changes in the transcriptome and the frequency of edited cells over time, showing that after an initial decline in the frequency of edited cells after transplant, they remained stable for several months. Up to 40% of peripheral blood T cells carried at least one edit, but there was a low frequency of cells carrying all three edited loci and the transgenic TCR, due to a lack of selection for these cells before transplant. The frequency of cells with the PDCD1 knockout, in particular, decreased over time, consistent with mouse studies indicating that these cells are less able to establish immunological memory, while cells with the TRAC/TRBC knockouts had transcriptional signatures of central memory in contrast to previous studies using only the knock-in of the transgenic TCR, which resulted in T cell exhaustion. This is an important first step in the development of next-generation immune cell products for personalized cell therapies.
Regulatory aspects of cell therapy
The European Medicines Agency (EMA) directive 2001/83/EC and Advanced Therapeutic Medicinal Products Regulation EC No 1394/2007 reads: ‘Somatic cell therapy medicinal product means a biological medicinal product which contains or consists of cells or tissues that have been subject to substantial manipulation so that biological characteristics, physiological functions or structural properties relevant for the intended clinical use have been altered, or of cells or tissues that are not intended to be used for the same essential function(s) in the recipient and the donor’. Similarly, the Food and Drug Administration (FDA), in Guidance for Human Somatic Cell Therapy and Gene Therapy – Guidance for Industry, March 1998, states that ‘Somatic cell therapy is the administration to humans of autologous, allogeneic, or xenogeneic living cells which have been manipulated or processed ex vivo [a particular topic’. This perhaps highlights the fact that], [due to its relative novelty]
Among the definition of ‘manipulations’ is genetic modification of the cells, and this implies that the regulations for gene therapy also have to be followed for cell therapy, as viral and non-viral vectors may be integral parts of the final product. Different territories are governed by different agencies, e.g. the Pharmaceutical and Food Safety Bureau (PFSB) in Japan, and the National Medical Products Administration (NMPA) in China, in addition to the FDA in the USA and the EMA in the EU. This results in different regulations regarding quality, safety, and efficacy for the same product. Thus, it is important to consider this when developing a pharmaceutical product - not a trivial task for small companies. Even the terminology can vary between agencies, and so a standardized medical terminology, MedDRA, has been developed by the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) – an organization that aims to bring together agencies and the pharmaceutical industry, providing guidelines to reduce differences between territories and facilitate sharing of regulatory information internationally. From a consumer point of view, i.e. the patient, the existence of different regulatory agencies and jurisdictions can make the difference between having access to a life-saving treatment or not. For this reason, any effort that aims to further align the regulatory agencies from different countries must be encouraged, promoted and supported. For example, the European Union (EU) has mutual recognition agreements (MRAs) of GMP inspections and batch certification of human medicines with countries including Australia, Canada, Japan and the US. This facilitates market access and international harmonization of standards, while reducing duplication of GMP inspections and costs for manufacturers. Unfortunately, cell therapy is excluded from the MRAs, therefore a product commercialized in one of the countries mentioned above will not be available in the EU unless the necessary inspections and batch testing are carried out by the EU, adding time and costs.
In cell therapy, the starting material consists of cells, most often obtained from human donors. This process requires strict adherence to the directives of the regulatory authorities, such as the obtaining of informed consent that must be provided by eligible donors and data protection that must be guaranteed by the manufacturer. An alternative scenario is represented by the case in which the starting material consists of a cell line. The regulations for the manufacturing of the cell line are expected to follow GMP regulations for clinical samples, but perhaps the most interesting aspect of this approach is related to the in vivo behavior of the cell line. Thorough pre-clinical studies must be carried out to demonstrate safety and efficacy. The first cell line to be used to provide the starting material for cell therapy is the cell line NK-92. This cell line, isolated from a patient with malignant non-Hodgkin’s lymphoma, has shown a high level of safety and efficacy in preclinical studies [92]Yan Y, Steinherz P, Klingemann HG et al. Antileukemia activity of a natural killer cell line against human leukemias. Clin. Cancer Res. 1998; 4: 2859–68., [93]Zhang J, Zheng H, Diao Y. Natural killer cells and current applications of chimeric antigen receptor-modified NK-92 cells in tumor immunotherapy. Int. J. Mol. Sci. 2019; 20., although the cells have to be irradiated before use because of their origin in lymphoma. NK-92 has been used as a starting material to manufacture the anti-HER2-CAR-CD28zeta-expressing allogeneic NK-92 cells, and anti-CD33 CAR NK cells, used in clinical trials NCT03383978 and NCT02944162 respectively (from clinicaltrials.gov). The data currently available indicate no issues with safety and no or mild reactions post-infusion, but unfortunately these products have not so far shown significant improvement in efficacy compared to the standard of care. Nevertheless, these clinical trials have paved the road to the use of cell lines as starting material, an approach that is central for the off-the-shelf model for cell therapy.
There are numerous challenges ahead for the development and manufacture of cell therapies, the most prominent being reduction of costs. This will be achieved through a combination of process optimization, automation, and the production of off-the-shelf alternatives. Innovation driven by the computation of large-scale data sets is expected to play a central part in the delivery of new solutions, ensuring quality and accessible products for a larger number of patients. Mogrify®, using its suite of platform technologies for direct cellular conversion and maintenance of cells, offers a transformative approach to the development of ’off-the-shelf’ cell therapies, reducing the processing time of cell differentiation, and reducing variability between batches, whilst at the same time increasing the scalability of the cell products and facilitating an entirely new class of in vivo reprograming therapies.
References
1. Delorme EJ, Alenxander P. Treatment of Primary Fibrosarcoma in the Rat with Immune Lymphocyte. Lancet 1964; 2: 117–20. Crossref
2. Rafiq S, Hackett CS, Brentjens RJ. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 2020; 17: 147–67. Crossref
3. Brocker T, Karjalainen K. Signals through T cell receptor-ζ chain alone are insufficient to prime resting T lymphocytes. J. Exp. Med. 1995; 181: 1653–9. Crossref
4. Maude SL, Laetsch TW, Buechner J et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018; 378: 439–48. Crossref
5. Schuster SJ, Svoboda J, Chong EA et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N. Engl. J. Med. 2017; 377: 2545–54. Crossref
6. Turtle CJ, Hanafi LA, Berger C et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 2016; 126: 2123–38. Crossref
7. Majzner RG, Mackall CL. Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med. 2019; 25: 1341–55. Crossref
8. Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020; 19: 185–99. Crossref
9. Rackham OJL, Firas J, Fang H et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016; 48: 331–5. Crossref
10. Schmidl C, Hansmann L, Lassmann T et al. The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood 2014; 123: 68–78. Crossref
11. Schmidl C, Renner K, Peter K et al. Transcription and enhancer profiling in human monocyte subsets. Blood 2014; 123: 90–9. Crossref
12. Wang M, Munoz J, Goy A et al. KTE-X19, an Anti-CD19 Chimeric Antigen Receptor (CAR) T Cell Therapy, in Patients (Pts) with Relapsed/Refractory Mantle Cell Lymphoma (R/R MCL): Results of the Phase 2 ZUMA-2 Study. Biol. Blood Marrow Transplant. 2020; 26: S1. Crossref
13. Riegler LL, Jones GP, Lee DW. Current approaches in the grading and management of cytokine release syndrome after chimeric antigen receptor T-cell therapy. Ther. Clin, Risk Manag. 2019; 15: 323–35. Crossref
14. Bonifant CL, Jackson HJ, Brentjens RJ, Curran KJ. Toxicity and management in CAR T-cell therapy. Mol. Ther. - Oncolytics 2016; 3: 16011. Crossref
15. Yu S, Yi M, Qin S, Wu K. Next generation chimeric antigen receptor T cells: Safety strategies to overcome toxicity. Mol. Cancer 2019; 18. Crossref
16. Giordano-Attianese G, Gainza P, Gray-Gaillard E et al. A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy. Nat. Biotechnol. 2020; 38: 426–38. Crossref
17. Kanda J. Effect of HLA mismatch on acute graft-versus-host disease. Int. J. Hematol. 2013; 98: 300–8. Crossref
18. Pegram HJ, Lee JC, Hayman EG et al. Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood 2012; 119: 4133–41. Crossref
19. Hu B, Ren J, Luo Y et al. Augmentation of Antitumor Immunity by Human and Mouse CAR T Cells Secreting IL-18. Cell Rep. 2017; 20: 3025–33. Crossref
20. Moon EK, Carpenito C, Sun J et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin. Cancer Res. 2011; 17: 4719–30. Crossref
21. Titov A, Valiullina A, Zmievskaya E et al. Advancing CAR T-cell therapy for solid tumors: Lessons learned from lymphoma treatment. Cancers (Basel) 2020; 12: 1–22. Crossref
22. Liu E, Marin D, Banerjee P et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020; 382: 545–53. Crossref
23. Rozenbaum M, Meir A, Aharony Y, Itzhaki O, Schachter J. Gamma-Delta CAR-T Cells Show CAR-Directed and Independent Activity Against Leukemia. Front. Immunol. 2020; 11: 1–8. Crossref
24. Klichinsky M, Ruella M, Shestova O et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020; 1–7. Crossref
25. Fang F, Xiao W, Tian Z. Challenges of NK cell-based immunotherapy in the new era. Front. Med. 2018; 12: 440–50. Crossref
26. Tonn T, Schwabe D, Klingemann HG et al. Treatment of patients with advanced cancer with the natural killer cell line NK-92. Cytotherapy 2013; 15: 1563–70. Crossref
27. Williams BA, Law AD, Routy B et al. A phase I trial of NK-92 cells for refractory hematological malignancies relapsing after autologous hematopoietic cell transplantation shows safety and evidence of efficacy. Oncotarget 2017; 8: 89256–68. Crossref
28. Zhang C, Burger MC, Jennewein L et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl Cancer Inst. 2016; 108: 1–12. Crossref
29. He Q, Jiang X, Zhou X, Weng J. Targeting cancers through TCR-peptide/MHC interactions. J. Hematol. Oncol. 2019; 12: 1–17. Crossref
30. Ping Y, Liu C, Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell 2018; 9: 254–66. Crossref
31. Levine BL, Miskin J, Wonnacott K, Keir C. Global Manufacturing of CAR T Cell Therapy. Mol. Ther. – Methods Clin. Dev. 2017; 4: 92–101. Crossref
32. Spink K, Steinsapir A. The long road to affordability: a cost of goods analysis for an autologous CAR-T process. Cell Gene Ther. Insights 2018; 4: 1105–16. Crossref
33. Brindley DA, Davie NL, Culme-Seymour EJ, Mason C, Smith DW, Rowley JA. Peak serum: Implications of serum supply for cell therapy manufacturing. Regen. Med. 2012; 7: 7–13. Crossref
34. Lanza R, Russell DW, Nagy A. Engineering universal cells that evade immune detection. Nat. Rev. Immunol. 2019; 19: 723–33. Crossref
35. Efremova M, Vento-Tormo R, Park J-E, Teichmann SA, James KR. Immunology in the Era of Single-Cell Technologies. Annu. Rev. Immunol. 2020; 38. Crossref
36. See P, Dutertre CA, Chen J et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science (80) 2017; 356. Crossref
37. Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science (80). 2017; 358: 58–63. Crossref
38. Yost KE, Chang HY, Satpathy AT. Tracking the immune response with single-cell genomics. Vaccine 2019; 38(28): 4487–90. Crossref
39. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018; 18: 35–45. Crossref
40. Method of the Year 2013. Nat. Methods. 2014; 11: 1. Crossref
41. Lareau CA, Duarte FM, Chew JG et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 2019; 37: 916–24. Crossref
42. Satpathy AT, Saligrama N, Buenrostro JD et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 2018; 24: 580–90. Crossref
43. Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015; 2015: 21.29.1–21.29.9. Crossref
44. Cusanovich DA, Daza R, Adey A et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science (80) 2015; 348: 910–4. Crossref
45. Bendall SC, Simonds EF, Qiu P et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science (80) 2011; 332: 687–96. Crossref
46. Packer J, Trapnell C. Single-Cell Multi-omics: An Engine for New Quantitative Models of Gene Regulation. Trends Genet. 2018; 34: 653–65. Crossref
47. Cao J, Cusanovich DA, Ramani V et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science (80) 2018; 361: 1380–5. Crossref
48. Parekh U, Wu Y, Zhao D et al. Mapping Cellular Reprogramming via Pooled Overexpression Screens with Paired Fitness and Single-Cell RNA-Sequencing Readout. Cell Syst. 2018; 7: 548–555.e8. Crossref
49. Datlinger P, Rendeiro AF, Schmidl C et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017. DOI:10.1038/nmeth.4177. Crossref
50. Rubin AJ, Parker KR, Satpathy AT et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 2019; 176: 361–376.e17. Crossref
51. Stoeckius M, Hafemeister C, Stephenson W et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017; 14: 865–8. Crossref
52. Mereu E, Lafzi A, Moutinho C et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 2020; 1–9. Crossref
53. Stuart T, Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019; 20: 257–72. Crossref
54. Paul F, Arkin Y, Giladi A et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 2015; 163: 1663–77. Crossref
55. Dutertre CA, Becht E, Irac SE et al. Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity 2019; 51: 573–589.e8. Crossref
56. Guilliams M, Dutertre CA, Scott CL et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity 2016; 45: 669–84. Crossref
57. Regev A, Teichmann SA, Lander ES et al. The human cell atlas. Elife 2017; 6. Crossref
58. Stubbington MJT, Lönnberg T, Proserpio V et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 2016; 13: 329–32. Crossref
59. Lindeman I, Emerton G, Mamanova L et al. BraCeR: B-cell-receptor reconstruction and clonality inference from single-cell RNA-seq. Nat. Methods 2018; 15: 563–5. Crossref
60. Setliff I, Shiakolas AR, Pilewski KA et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell 2019; 179: 1636–1646.e15. Crossref
61. Mimitou EP, Cheng A, Montalbano A et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 2019; 16: 409–12. Crossref
62. Watcham S, Kucinski I, Gottgens B. New insights into hematopoietic differentiation landscapes from single-cell RNA sequencing. Blood 2019; 133: 1415–26. Crossref
63. Pijuan-Sala B, Guibentif C, Göttgens B. Single-cell transcriptional profiling: A window into embryonic cell-type specification. Nat. Rev. Mol. Cell Biol. 2018; 19: 399–412. Crossref
64. Trapnell C. Defining cell types and states with single-cell genomics. Genome Res. 2015; 25: 1491–8. Crossref
65. La Manno G, Soldatov R, Zeisel A et al. RNA velocity of single cells. Nature 2018; 560: 494–8. Crossref
66. Trapnell C, Cacchiarelli D, Grimsby J et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32: 381–6. Crossref
67. Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 2016; 13: 845–8. Crossref
68. Li Y, Hermanson DL, Moriarity BS, Kaufman DS. Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell Stem Cell 2018; 23: 181-192.e5. Crossref
69. Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987; 51: 987–1000. Crossref
70. Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006; 126: 663–76. Crossref
71. Rosa FF, Pires CF, Kurochkin I et al. Direct reprogramming of fibroblasts into antigen-presenting dendritic cells. Sci. Immunol. 2018; 3: 1–16. Crossref
72. Duan J, Li B, Bhakta M et al. Rational Reprogramming of Cellular States by Combinatorial Perturbation. Cell Rep. 2019; 27: 3486-3499.e6. Crossref
73. Morris SA, Cahan P, Li H et al. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 2014; 158: 889–902. Crossref
74. Kamaraj US, Gough J, Polo JM, Petretto E, Rackham OJL. Computational methods for direct cell conversion. Cell Cycle 2016; 15: 3343–54. Crossref
75. D’Alessio AC, Fan ZP, Wert KJ et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015; 5: 763–75. Crossref
76. Okawa S, Saltó C, Ravichandran S et al. Transcriptional synergy as an emergent property defining cell subpopulation identity enables population shift. Nat. Commun. 2018; 9: 1–10. Crossref
77. Park JE, Botting RA, Conde CD et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (80) 2020; 367: eaay3224. Crossref
78. Themeli M, Rivière I, Sadelain M. New cell sources for T cell engineering and adoptive immunotherapy. Cell Stem Cell 2015; 16: 357–66. Crossref
79. Minagawa A, Yoshikawa T, Yasukawa M et al. Enhancing T Cell Receptor Stability in Rejuvenated iPSC-Derived T Cells Improves Their Use in Cancer Immunotherapy. Cell Stem Cell 2018; 23: 850-858.e4. Crossref
80. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc 2020; 1–23. Crossref
81. Yoosuf N, Navarro JF, Salmén F, Ståhl PL, Daub CO. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res 2020; 22: 1–10. Crossref
82. Berglund E, Maaskola J, Schultz N et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018; 9. Crossref
83. Yost KE, Satpathy AT, Wells DK et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019; 25: 1251–9. Crossref
84. Satpathy AT, Granja JM, Yost KE et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019; 37: 925–36. Crossref
85. Lynn RC, Weber EW, Sotillo E et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019; 576: 293–300. Crossref
86. Dixit A, Parnas O, Li B et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016; 167: 1853-1866.e17. Crossref
87. Dong MB, Wang G, Chow RD et al. Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell 2019; 178: 1189–1204.e23. Crossref
88. Wei J, Long L, Zheng W et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 2019. Crossref
89. Ye L, Park JJ, Dong MB et al. In vivo CRISPR screening in CD8 T cells with AAV–Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotechnol. 2019; 37: 1302–13. Crossref
90. Roth TL, Li PJ, Blaeschke F et al. Pooled Knockin Targeting for Genome Engineering of Cellular Immunotherapies. Cell 2020; 181: 728–744.e21. Crossref
91. Stadtmauer EA, Fraietta JA, Davis MM et al. CRISPR-engineered T cells in patients with refractory cancer. Science (80- ) 2020; 367: eaba7365. Crossref
92. Yan Y, Steinherz P, Klingemann HG et al. Antileukemia activity of a natural killer cell line against human leukemias. Clin. Cancer Res. 1998; 4: 2859–68. Crossref
93. Zhang J, Zheng H, Diao Y. Natural killer cells and current applications of chimeric antigen receptor-modified NK-92 cells in tumor immunotherapy. Int. J. Mol. Sci. 2019; 20. Crossref
Affiliations
Vicki Moignard
Mogrify Limited, 25 Cambridge Science Park, Milton Road, Cambridge, CB4 0FW, UK
Alessandra De Riva
Mogrify Limited, 25 Cambridge Science Park, Milton Road, Cambridge, CB4 0FW, UK
Raul Elgueta
Mogrify Limited, 25 Cambridge Science Park, Milton Road, Cambridge, CB4 0FW, UK
Authorship & Conflict of Interest
Contributions: All named authors take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Acknowledgements: None.
Disclosure and potential conflicts of interest: The authors declare that they have no conflicts of interest.
Funding declaration: The authors received no financial support for the research, authorship and/or publication of this article.
Article & copyright information
Copyright: Published by Cell and Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0 which allows anyone to copy, distribute, and transmit the article provided it is properly attributed in the manner specified below. No commercial use without permission.
Attribution: Copyright © 2020 Moignard V, De Riva A & Elgueta R. Published by Cell and Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0.
Article source: Invited.
Revised manuscript received: Oct 26 2020; Publication date: Oct 29 2020.

.jpg)