Automated plasmid assembly: a perspective from early drug discovery
Nucleic Acid Insights 2024; 1(10), 363–370
DOI: 10.18609/nai.2024.045
Plasmid reagent generation is often both the first and the rate limiting step in early drug discovery projects. At AstraZeneca, we have sought to accelerate the plasmid DNA generation process by integrating laboratory automation and in silico processes from digital sequence to clonal plasmid construct. These processes are tracked in a custom-built application, called the Construct Request Portal (CRP), which also hosts various bioinformatic applications such as bespoke codon optimization algorithms and analysis of sequencing data by Oxford Nanopore Technologies (ONT). Drug discovery activities are diverse and may range from requiring a single cell-line engineering construct to variant libraries containing thousands of plasmid variants. Ultimately, the throughput, length, and complexity of plasmid constructs will trigger different DNA fragment formats and assembly pipelines. Herein, we will review the field and discuss advantages and disadvantages of ‘do-it-yourself’ (DIY) plasmid DNA assembly workflows in the perspective of workflow requirements and rapid sourcing of synthetic DNA.
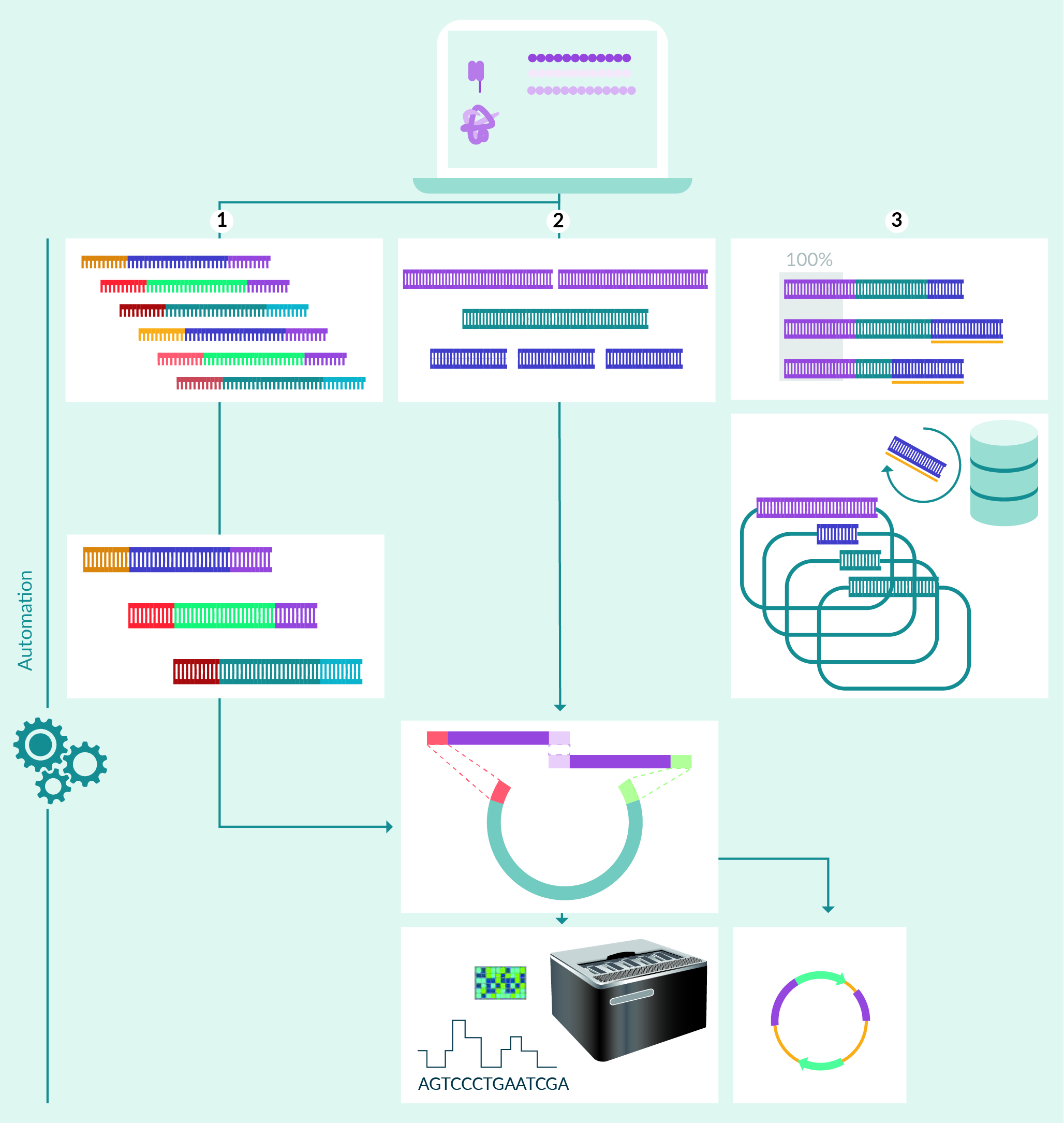
Synthetic plasmid DNA assembly workflows. Amino acids are entered into the Construct request portal (CRP) which codon optimizes and fragments the DNA sequences in sizes suitable for one of three cloning workflows: (1) Gene assembly from arrayed or pooled oligos; (2) Gene assembly from dsDNA fragments; (3) Gene assembly from clonal DNA fragments.
