Technical transfer of a rapid microbial platform for vaccine production
Vaccine Insights 2023; 2(4), 133–146
DOI: 10.18609/vac.2023.024
The limited availability and affordability of vaccines to low- and middle-income countries (LMIC) has created a need for solutions that will ensure effective, affordable vaccine production technology. To establish a rapid and economical platform for the expression of viral proteins in high yield and purity by Pichia pastoris (X33), the receptor-binding domain (RBD) protein of the SARS-CoV2 was selected in this study. After fermentation at the 5 L scale, the protein was purified by a simplified chromatography, with minimal sample treatment. The purified protein was characterized biochemically, and after its formulation, the immunogenicity was evaluated in mice. Collectively, the data suggested that the vaccine candidate is a suitable COVID-19 vaccine candidate antigen for technology transfer. Furthermore, this study creates a robust foundation for industrial production at scale.
Vaccines are one of the greatest medical advances of modern times, bringing immense value to children, families, communities, and economies. However, not every country has the technology and infrastructure to develop, test and manufacture new vaccines. Therefore, many countries must rely on others for vaccine supplies to save lives when a pandemic strikes [1]Choi EM. COVID-19 Vaccines for Low- And Middle-Income Countries. Trans. R. Soc. Trop. Med. Hyg. 2021, 115; 447–456. [2]Mihigo R, Okeibunor J, Cernuschi T, Petu A, Satoulou A, Zawaira F. Improving Access to Affordable Vaccines for Middle-Income Countries in the African Region. Vaccine 2019, 37, 2838–2842..
The advent of COVID-19 accelerated the understanding of the need for research and rapid manufacture of vaccines and LMIC have been working towards becoming self-sufficient in vaccine production. In this study, we report on research carried out in collaboration with PT Bio Farma as part of a feasibility study funded by Future Vaccine Manufacturing Research Hub (Vax-Hub) [3]Engineering UD of B. The Future Vaccine Manufacturing Research Hub: Securing the Supply of Essential Vaccines. (Accessed on 29 March 2023). to undertake technology transfer for the manufacture of RBD Spike SARS-CoV2 [4]Lan J, Ge J, Yu J et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. , using a microbial platform.
Over 30% of all biologics are produced in cell engineered microbial factories. Amongst the host cells available Pichia pastoris (Komagataella phaffii) has shown great potential and is currently reported in 13 clinical trials with more than 70 products already on the market or in late-stage
development [5]de Sá Magalhães S, Keshavarz‐Moore EP. Pastoris (Komagataella Phaffii) as a Cost‐effective Tool for Vaccine Production for Low‐ and Middle‐income Countries (Lmics). Bioeng. 2021, 8. [6]Kriesi WPT. by Pichia Produced Products on the Market. (Accessed on 29 March 2023)..
This study addresses integrated upstream and downstream operations, aiming to create an easy-to-scale process, emphasizing key analytics and the need for a robust, simple platform.
Methodology
Cells & seed cultures
Design, selection, and characterization of the cells and seed culture were performed by the partner company, PT Bio Farma. The selected gene of interest was cloned into the Pichia expression vector pPICZa-A, followed by the transformation of Komagataella phaffii (X-33, Mut+), selection, and confirmation steps to evaluate the clones as described below:
1. Transformation Escherichia coli (E. coli)TOP10 with pPICZα-A _RBD Spike SARSCOV2
E. coli TOP10 was transformed using 40 ng of plasmid pPICZa-A _RBD Spike SARSCOV2 by CaCl2 method (RBD Spike SARSCOV2 cloning sequence is PT Bio Farma proprietary and therefore cannot be disclosed). Antibiotics Zeocin™ 25 µg/mL and tetracycline 10 µg/mL were used for screening transformant E. coli TOP10/ pPICZa-A _RBD Spike SARSCOV2 in Luria Bertani media. Furthermore, the plasmid was isolated from transformant E. coli TOP10/ pPICZa-A _RBD Spike SARSCOV2 using Qiaprep Spin Miniprep Kit (50) (Cat: 27104), then verified by restricted analysis.
2. Transformation P. pastoris X-33 with pPICZα-A _RBD Spike SARSCOV2
Plasmid pPICZα-A _RBD Spike SARSCOV2was digested by enzyme SacI (NEB R0156S), then the linearised plasmid was purified using QIAquick Gel Extraction Kit (50) (Cat: 28704). P. pastoris X33 (his4) was transformed with 5.4 µg total DNA concentration of linearized plasmid by electroporation method (1727 V, 2.2 ms, electroporation cuvette 0.2 cm). Transformants X33/pPICZα-A _RBD Spike SARSCOV2were selected on yeast peptone dextrose (YPD) with 500 µg/mL and analyzed for methanol utilization (Mut+) using minimal methanol media and MD media. Selected transformants were cultured in YPD broth. As PCR template, genomes of transformants were isolated using MasterPureTM Yeast DNA Purification Kit (Cat: MPY80200) and then confirmed for homologous recombination of expression cassette RBD to P. pastoris genome using PCR analysis with AOX1 primers according to EasySelectTM Pichia Expression Kit Manual. The expression cassette RBD was confirmed through the appearance of a 1206 bp molecular weight band in agarose gel analysis.
3. Generation of research cell bank
A. Cell bank 1
One colony of transformant X33/ pPICZα-A _RBD Spike SARSCOV2 #2C was inoculated to 5 mL YPD media with 500 µg/mL Zeocin in a 50 mL conical tube. The culture was grown at 30 °C for 17 h with optical density (OD)600 of 8.6. Subsequently, glycerol was added to the culture to a final volume of 15%, then 1 mL was aliquoted into the cryovial, labeled, and stored at –80 °C.
B. Cell bank 2
500 L of P. pastoris X33/pPICZa-A _RBD Spike SARSCOV2 #2C was inoculated in 50 mL YPD media with 500 µg/mL Zeocin in a 1 L baffled flask. The culture was grown at 30°C for 18 h to an OD600 of 2.7. The culture was concentrated using centrifugation at 3000 xg for 5 min, to a final OD600 of 6.4. Furthermore, glycerol was added to the culture at a final concentration of 15%. Then 1 mL was aliquoted into the cryovial and stored at –80 °C and labeled.
C. Generation of research cell bank
500 µL of the seed, P. pastoris X33/ pPICZa-A _RBD Spike SARSCOV2 #2C was inoculated in 200 mL of buffered minimal glycerol media in a 1L baffled flask. The culture was grown at 30 °C for 28 h. After 28 h, glycerol was added to the culture at a final concentration of 20%. Afterward, it was aliquoted 1 mL into cryovials and stored at –80 °C as the research cell bank.
Fermentation process
Batch culture
Fermentations were carried out following Invitrogen’s protocol for Mut+ cells (Invitrogen, Pichia fermentation process guidelines. 2002 (version B 053002) [7]Invitrogen Corporation Pichia Fermentation Process Guidelines Overview Overview, Continued. Prog. Bot. 2002, 67, 1–11. with some modifications, and using an Ambr® 250 modular microbial system (Sartorius Stedim Biotech, Royston, UK). The bioreactors and single-use vessels were aseptically filled with sterile media and feed solutions. The initial fermentation volume was 100 mL of basal salt medium with 0.65 mL of PTM1 trace salts. The operating conditions were 28 °C, 30% dissolved oxygen (DO; using air or a mix of air and oxygen when required at 0.5 vvm), pH 5.00 ± 0.15 (controlled with 10% [v/v] ammonium hydroxide), and antifoam (polypropylene glycol 2000) was automatically added by the system when required. Reactors were inoculated via the septum to an OD600 of 1.
Measurements from off-gas CO2 sensors in the Ambr 250 were used by the system to calculate the carbon evolution rate (CER). The observation of a DO spike and a 20% drop in CER indicated the end of the batch phase.
Fed-batch culture optimization
Glycerol feed started automatically at a rate of 18.15 mL/h per liter of the initial fermentation volume (Li Vol), followed by a methanol adaptation phase using a feed rate of 3.6 mL/h//Li Vol. At this point, the combination of a design of experiments (DoE) approach with use of the Ambr250 microbioreactor was explored. Process setpoints and conditions for the design were imported into the Ambr 250 software via a DoE tag interface. Temperature, pH, %DO, and methanol feed were manipulated as factors.
A decrease in CER during methanol adaptation indicated depletion of glycerol and was used as a signal by the system to increase methanol feed rate, using an incremental step profile programmed into the Ambr 250 software (7.3 mL/h/Li Vol for 2 h and 10.9 mL/h/ Li Vol for the remainder of the fermentation). Glycerol and methanol feed rates were those specified by Invitrogen’s protocol for Mut+ cells.
At the end of fermentation, the secreted product was collected. The cell broth was centrifuged at 10,000 × g for 15 min and the supernatant was filtered using 0.80 µm cellulose nitrate and 0.45 µm polyvinylidene difluoride membrane filters (GE Healthcare Life Sciences, Buckinghamshire, UK). Samples were aliquoted into 30 mL and stored at –20 °C until further analysis.
RBD purification platform
A chromatography study was performed on an AKTA Avant system (GE Healthcare Life Sciences), equipped with a 2 mm path length UV cell with variable wavelength UV detection, measuring at three fixed wavelengths of 280, 260, and 320 nm. Chromatography parameters were monitored and controlled using UNICORN 6.2 software (GE Healthcare Life Sciences). Samples were filtered with a 0.45 µm polyethersulfone (PES) filter before loading.
Affinity chromatography
Samples were purified using an AVIPure®–COV2S Gen2 resin in a 1 mL column. Briefly, the column was equilibrated with 2 column volume (CV) of 25 mM N-(2-Hydroxy ethyl)-piperazine ethane sulfonic acid (HEPES), pH=7.4, and 20mL of sample was injected at a flow rate of 1mL/min. The column was washed with 4 CV 25mM HEPES, pH=7.4 until stable UV absorbance and conductivity were achieved. Two more washing steps were performed using 2 CV 0.5 M Tris, 25 mM HEPES, pH=7–9, and 3 CV of the buffer used for wash one. A linear gradient of 0–100% with 5 CVs of 1M Arginine-HCl, 50mM HEPES, pH=9 was used for elution at 1 mL/min. Both flowthrough and elution peaks were collected in fractions of 1.5 mL and stored at 4 °C for further analysis. A cleaning-in-place procedure was performed using 0.1 M NaOH, 1 M NaCl at 1 mL/min (4 CVs).
Analytics for product characterization
Biochemical assays
Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE samples were mixed with 4X SDS-PAGE sample loading buffer before being boiled at 95°C for 5 min. The 4X SDS-PAGE sample loading buffer contains 50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 1% b-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue. Samples were then applied (1 µg per well) to the lanes of a NuPAGE 4-12% Bis-Tris gel (Thermo Fisher Scientific) at 200 V for 45 min. The gels were then stained with InstantBlueTM Coomassie Protein Stain and imaged using the GE AmershamTM Imager 600 (Pittsburgh, PA, USA).
Western blot (WB)
For WB, SDS-PAGE was performed as above, but the gels were not stained. Instead, the gels were transferred to the Trans-Blot Turbo Transfer Pack membrane using the Bio-Rad Trans-Blot® TurboTM Transfer System (Hercules, CA, USA). The membrane, 0.2 µm nitrocellulose, was blocked in phosphate-buffered saline (PBS) with Tween 20 (T) and 3% bovine serum albumin (BSA) for 45 min. The membrane was incubated with the primary antibody (Sino Biological Europe GmbH [Europe]) diluted 1:1000 in PBST-BSA for 2 h. The membrane was washed with PBST three times before being incubated with the secondary antibody (Cayman Chemical, USA), an anti-rabbit horseradish peroxidase (HRP)-conjugated antibody, diluted 1:1,000 in PBST-BSA for 1 h. The membrane was washed with PBST twice and PBS once. The membrane was incubated with the Thermo Fisher Scientific (Paisley, UK) Pierce ECL Western Blotting Substrate for 1–2 min in darkness then exposed and imaged using the GE AmershamTM Imager 600.
Dot blot
2 µL of each sample was applied as a ‘dot’ on an 8×8 mm, 0.2 µm nitrocellulose membrane. Samples were measured in triplicate. The membrane was incubated, washed, and imaged in the same fashion as the WB membranes.
Animal studies
Vaccination
Groups of BALB/c mice were vaccinated (n=7 mice) with 200 µg adjuvant alum (alhydrogel 1.3% [Croda, UK]) only, 2.5 µg, 5 µg, or 10 µg RBD antigen formulated with 200 µg adjuvant alum or in combination with 20 µg CPG adjuvant, which is a synthetic immunostimulatory oligonucleotide that contains unmethylated CpG dinucleotides and a Toll-like receptor 9-agonist (Invivogen, USA), or with complete freund’s adjuvant (Merck, Germany) (1:1) on day 0 and again on day 20. Vaccination was administered subcutaneously at the base of the tail at a total volume of 200 µL/dose. Sera were collected from blood taken 2 weeks after the second inoculation.
Antibody titer determination by ELISA
Flat-bottom-well polyvinyl plates (96 wells/plate) were coated with RBD antigen (2.5 µg/mL) in PBS for 18–20 h at room temperature in a humidified atmosphere and left overnight. Unbound antigen was discarded and BSA (10 mg/mL) in PBS was added as a blocking solution for 1 h before washing with PBS containing v/v 0.05% Tween-20. Serial dilutions (log 1,5 to log 5,0) of sera obtained from immunized mice were added to wells and held overnight at room temperature. After washing, antigen-bound antibody was detected using HRP-conjugated goat anti-mouse antibodies or for 1 h at room temperature. 100 µL of 3,3′,5,5′-tetramethylbenzidine (TMB) was then added and left to develop for 15 min before the addition of 2 M H2SO4 for 50 µL/well to stop the reaction. Antibody titers were expressed as the reciprocal of the highest dilution of serum required to achieve an OD of 0.2.
Neutralization test
Neutralization testing methodology was as per the protocol from SARS-CoV-2 Surrogate Virus Neutralization Test (sVNT) manufacturer (Genscript). Samples and positive or negative controls were mixed at a 1:1 (v/v) ratio with HRP-conjugated RBD and incubated at 37 °C for 30 min. A total of 100 µL of each sample, positive control, or negative control were placed on the sVNT plate and incubated for 30 min at 37 °C. The plate was washed four times with 1x wash buffer. 100 µL of TMB was added to the plate, which was then incubated in the dark for 15 min at room temperature (20–25 °C). To terminate the reaction, 50 µL of stop solution was added to the plate, and the absorbance of the sample was measured at 450 nm.
Results
The key technical outputs achieved in this study comprise the determination of fermentation conditions for a recombinant COVID-19 vaccine, the development of a simplified purification protocol for a COVID-19 vaccine antigen, and the scalability of a low-cost process for a COVID-19 vaccine.
These achievements are summarized in four phases as described below.
Phase 1: the construction of a recombinant SARS-CoV-2 RBD of spike protein expressed in
P. pastoris as a vaccine antigen
The first phase of this study consisted of the construction of a recombinant SARS-CoV-2 RBD of spike (S) protein expressed in P. pastoris as a vaccine antigen. The merit for selection of RBD as a sole vaccine immunogen relies on the reliable elicitation of neutralizing antibodies that target RBD following natural infection or vaccination, a key biomarker of protection [8]Kleanthous H, Silverman JM, Makar KW, Yoon IK, Jackson N, Vaughn DW. Scientific Rationale for Developing Potent RBD-Based Vaccines Targeting COVID-19. npj Vaccines 2021, 6, 1–10.. Moreover, the potential manufacturing and cost advantages of using RBD as a vaccine immunogen across different platform technologies as well as the engagement of experienced developing country vaccine manufacturers as partners in delivering billions of vaccines doses, makes RBD vaccine a promising candidate to address a critical medical need and ensure equitable access of vaccine [9]Lee J, Liu Z, Chen WH et al. Process Development and Scale-up Optimization of the SARS-CoV-2 Receptor Binding Domain–Based Vaccine Candidate, RBD219-N1C1. Appl. Microbiol. Biotechnol. 2021, 105, 4153–4165. [10]Dalvie NC, Biedermann AM, Rodriguez-Aponte SA et al. Scalable, Methanol-Free Manufacturing of the SARS-CoV-2 Receptor-Binding Domain in Engineered Komagataella Phaffii. Biotechnol. Bioeng. 2022, 119, 657–662..
The clone was designed and selected by the partner company (Figures 1A & 1B). The results of the study showed that the RBD clone was constructed successfully using vector pPICZ alpha with AOX1 promoter in P. pastoris and RBD protein was expressed extracellularly as shown in the WB analysis (Figure 1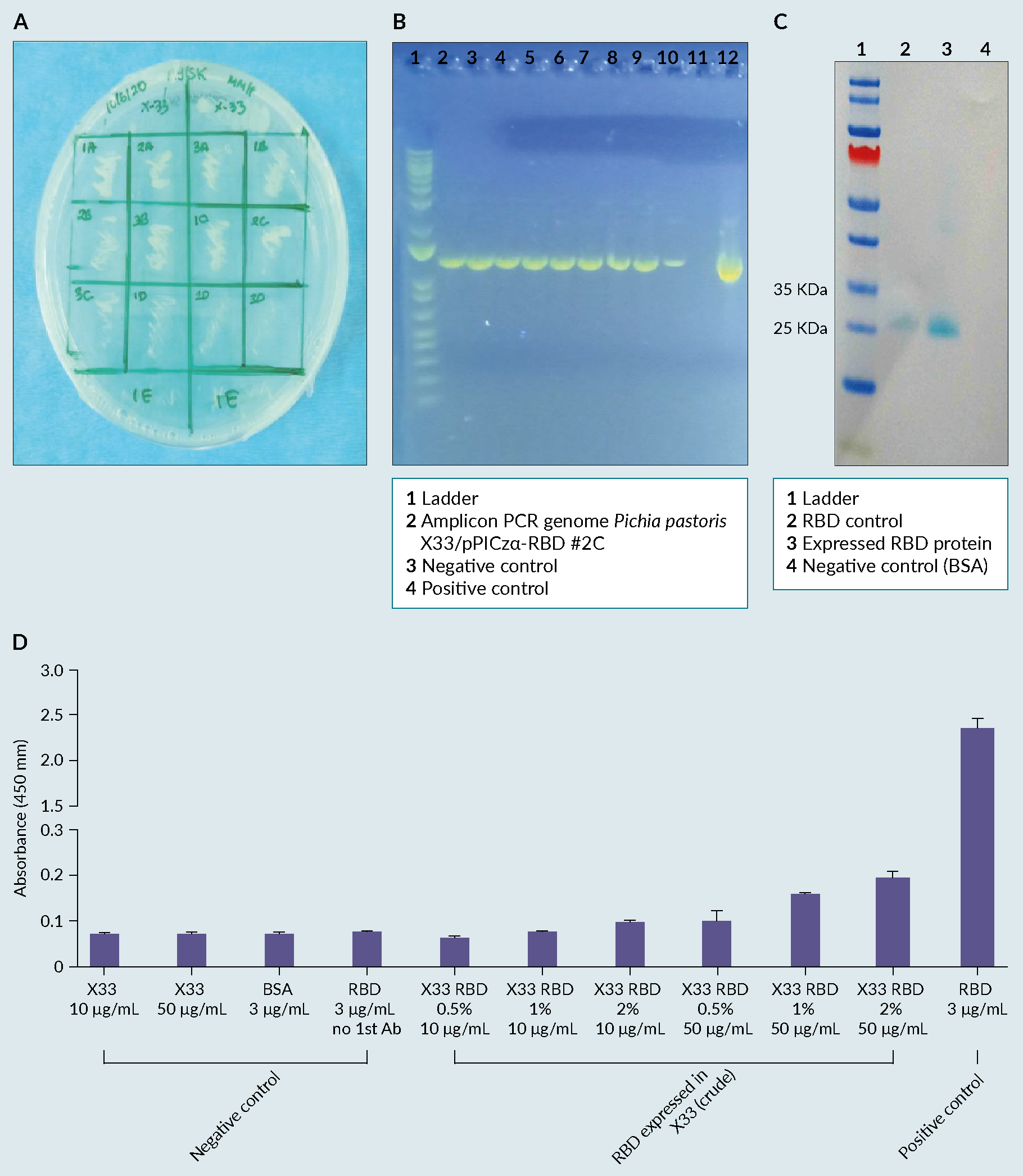 A) Clone selection using Methanol Phenotype test, in Minimal Methanol Histidine (MMH) left side; B) PCR analysis of Pichia integrands with AOX1 primers according to EasySelectTM Pichia Expression Kit , band with molecular weight of 1206 bp; C) Biochemical evaluation of the expressed RBD protein, using a western blot analysis, where the band of interested is shown on lane 2-10; D) Evaluation of the biological activity of the expressed RBD protein, using a commercially available ELISA, where the highest absorbance was obtained for: X33 RBD 2% 50 μg/mL.C), with a weight of ~23 kDa. Its biological activity was then confirmed by reactivity of the antigen to commercially available anti-SARS-CoV-2 RBD antibody by ELISA (Figure 1D).
A) Clone selection using Methanol Phenotype test, in Minimal Methanol Histidine (MMH) left side; B) PCR analysis of Pichia integrands with AOX1 primers according to EasySelectTM Pichia Expression Kit , band with molecular weight of 1206 bp; C) Biochemical evaluation of the expressed RBD protein, using a western blot analysis, where the band of interested is shown on lane 2-10; D) Evaluation of the biological activity of the expressed RBD protein, using a commercially available ELISA, where the highest absorbance was obtained for: X33 RBD 2% 50 μg/mL.C), with a weight of ~23 kDa. Its biological activity was then confirmed by reactivity of the antigen to commercially available anti-SARS-CoV-2 RBD antibody by ELISA (Figure 1D).
Phase 2: the development of a small-scale fermentation process using a DoE approach
To improve the production process of the antigen, the evaluation of the key parameters of fermentation processes was performed on small-scale bioreactors using a DoE approach. Process and production systems where physical, chemical, or biological transformations take place can benefit substantially by using DoE as a design and optimization tool [11]Bowden GD, Pichler BJ, Maurer A. A Design of Experiments (DoE) Approach Accelerates the Optimization of Copper-Mediated 18F-Fluorination Reactions of Arylstannanes. Sci. Rep. 2019, 9, 1–10.. The DoE methodology is particularly useful for avoiding experimental biases and significantly reducing the required number of experiments. In this study, the method applied was a Box–Behnken design [12]Peng X, Yang G, Shi Y, Zhou Y, Zhang M, Li S. Box–Behnken Design Based Statistical Modeling for the Extraction and Physicochemical Properties of Pectin from Sunflower Heads and the Comparison with Commercial Low-Methoxyl Pectin. Sci. Rep. 2020, 10, 1–10., where four factors (temperature [°C], pH, %DO, and methanol feed) with three levels each were evaluated, leading to a total of 28 experiments (Table 1). Optimizing the production process of RBD gives a significant impact on manufacturing efficiency and productivity and hence manufacturing cost in the long run.
| Table 1 Box-Behnken experimental design and response values for production yield. A) three levels (-1, 0 and 1) of factor X1 (Temperature) represented 26, 28 and 30 °C, respectively. B) three levels (-1, 0 and 1) of factor X2 (pH) represented 3.5, 5.0 and 6.5, respectively. C) three levels (-1, 0 and 1) of factor X3 (% dissolved oxygen) represented 20, 30 and 40%, respectively. D) three levels (-1, 0 and 1) of factor X4 (% Methanol) represented 8.9, 10.9 and 12.9. | |||||
| Exp No. | CODES | % RBD content | |||
| X1a | X2b | X3c | X4d | ||
| 1 | -1 | -1 | 0 | 0 | 12.69 |
| 2 | -1 | -1 | 0 | 0 | 0.04 |
| 3 | 1 | 0 | 0 | 0 | 5.91 |
| 4 | 0 | 0 | 0 | 0 | 1.46 |
| 5 | 0 | 0 | -1 | -1 | 1.33 |
| 6 | 0 | 0 | -1 | 1 | 0.12 |
| 7 | 0 | 0 | 1 | -1 | 1.10 |
| 8 | 0 | 0 | 1 | 1 | 0.24 |
| 9 | -1 | 0 | 0 | -1 | 3.73 |
| 10 | -1 | 0 | 0 | 1 | 2.27 |
| 11 | 1 | 0 | 0 | -1 | 0.04 |
| 12 | 0 | 0 | 0 | 0 | 0.73 |
| 13 | 0 | -1 | -1 | 0 | 9.00 |
| 14 | 0 | -1 | 1 | 0 | 11.03 |
| 15 | 0 | 1 | -1 | 0 | 12.54 |
| 16 | 0 | 1 | 1 | 0 | 1.87 |
| 17 | -1 | 0 | -1 | 0 | 6.83 |
| 18 | -1 | 0 | 1 | 0 | 4.36 |
| 19 | 0 | 0 | 0 | 0 | 0.59 |
| 20 | 1 | 0 | -1 | 0 | 0.06 |
| 21 | 0 | -1 | 0 | -1 | 10.69 |
| 22 | 0 | -1 | 0 | 1 | 15.57 |
| 23 | 0 | 1 | 0 | -1 | 0.53 |
| 24 | 0 | 1 | 0 | 1 | 17.66 |
| 25 | 1 | 1 | 0 | 0 | 25.52 |
| 26 | 0 | 0 | 0 | 0 | 1.23 |
| 27 | 1 | 0 | 1 | 0 | 0.06 |
| 28 | 1 | 0 | 0 | 1 | 0.06 |
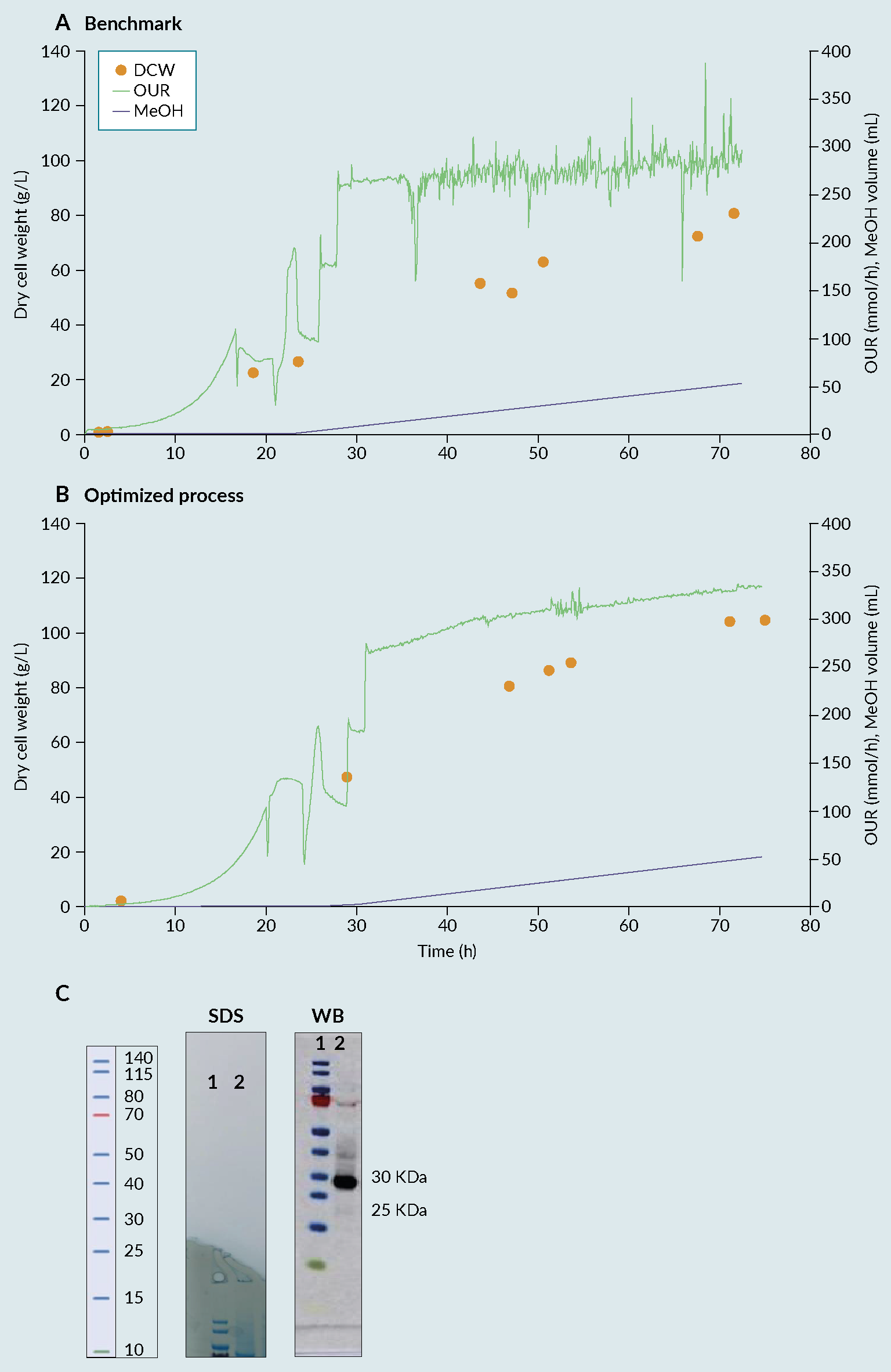
By combining Ambr 250 experimentation with statistical DoE, an improved process was identified, enabling a 30% increase in biomass compared with the benchmark (Invitrogen protocol) (Figure 2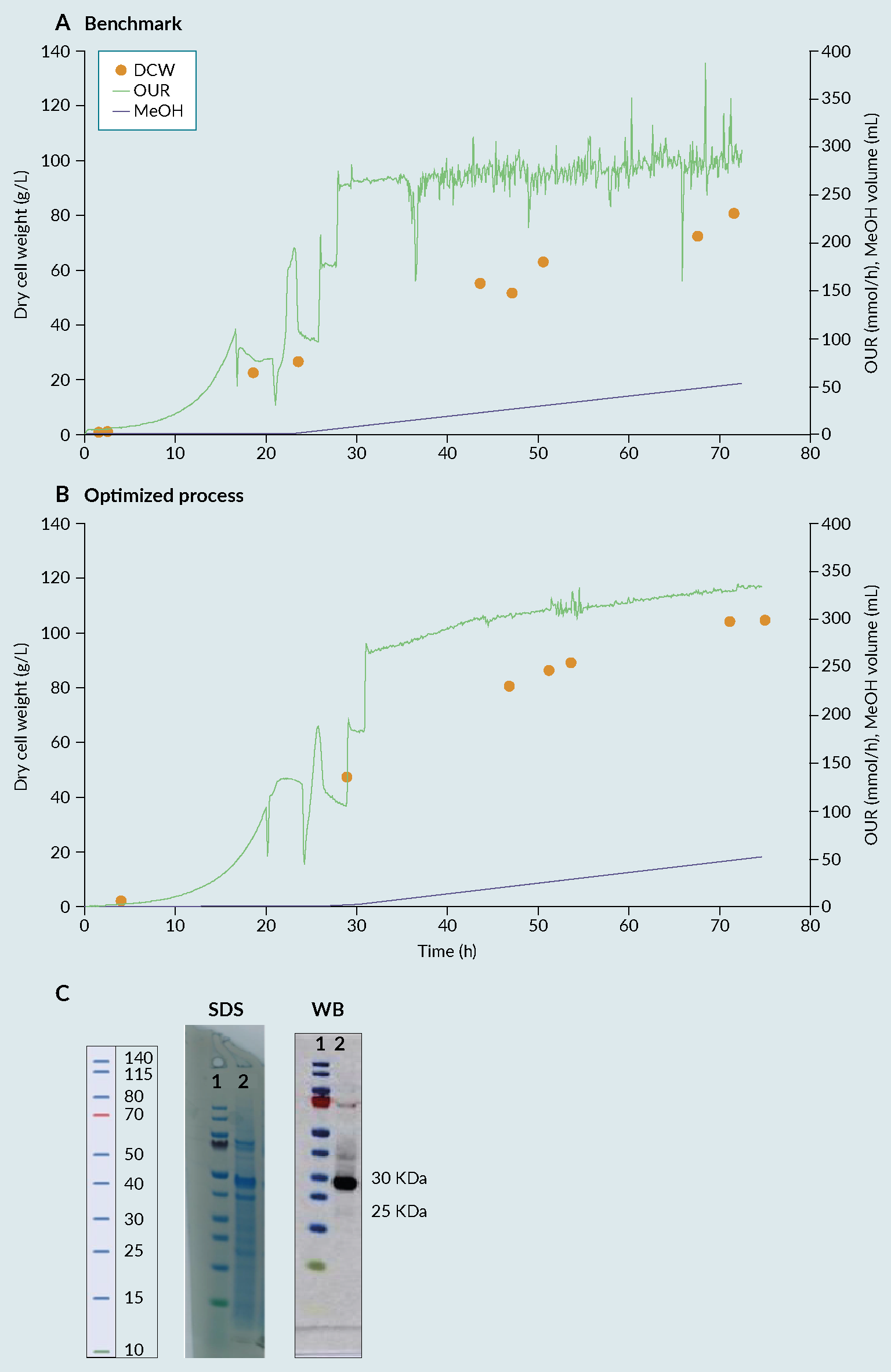 Fermentation profile of cultivation process: (A) benchmark, (B) optimised and (C) biochemical evaluation via SDS-PAGE (SDS) and Western Blot (WB) of produced receptor-binding domain (RBD) from optimised process. Lane 1 representing the protein Ladder and Lane 2 representing the produced RBD with a size between 25 and 30kDa.) [7]Invitrogen Corporation Pichia Fermentation Process Guidelines Overview Overview, Continued. Prog. Bot. 2002, 67, 1–11.. The 28 samples were then analyzed by dot blot and compared to a standard curve using a commercial RBD Ag. The relative titers were then quantified based on intensities using ImageQuant software (Figure 3
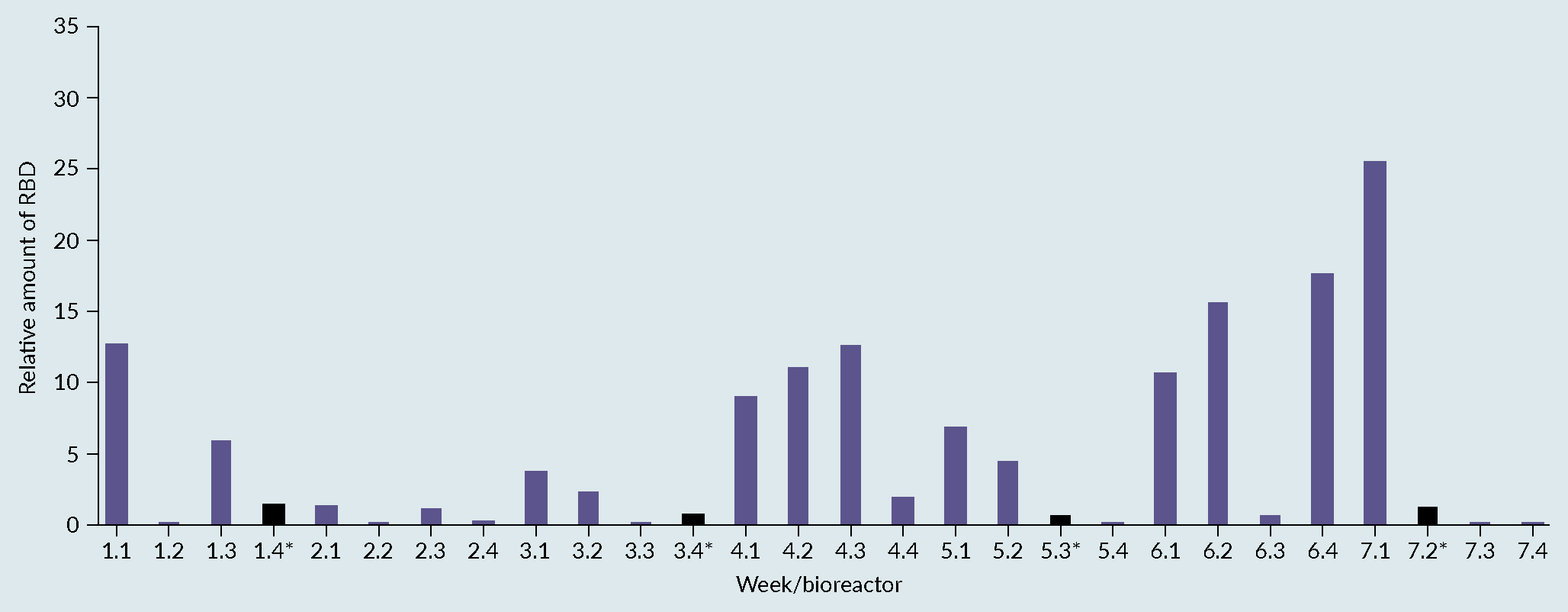
Fermentation profile of cultivation process: (A) benchmark, (B) optimised and (C) biochemical evaluation via SDS-PAGE (SDS) and Western Blot (WB) of produced receptor-binding domain (RBD) from optimised process. Lane 1 representing the protein Ladder and Lane 2 representing the produced RBD with a size between 25 and 30kDa.) [7]Invitrogen Corporation Pichia Fermentation Process Guidelines Overview Overview, Continued. Prog. Bot. 2002, 67, 1–11.. The 28 samples were then analyzed by dot blot and compared to a standard curve using a commercial RBD Ag. The relative titers were then quantified based on intensities using ImageQuant software (Figure 3 Densitometry analysis of the 28 runs. Each bar in the graphic represents the relative amount of RBD obtained on each bioreactor using a certain set of cultivation parameters. The quantification was performed using a dot blot and the images analyzed using ImageQuant software (densitometry). The relative amount was then calculated based on a standard curve and normalized towards the process control (central point) to facilitate the evaluation of the effect of varying the factors considered. (*) process control, representing a set of parameters that, based on the literature and previous experiments, were known to lead to the production of RBD.). From the figure, reactor 1 from week 7 (7.1), gave the best relative percentage, around 25% in comparison with the process control. As a result of the good reproducibility, regardless of run week, batch variability or position in the Ambr 250 module, the results from the full experimental design can be attributed to changes in factor levels with confidence.
Densitometry analysis of the 28 runs. Each bar in the graphic represents the relative amount of RBD obtained on each bioreactor using a certain set of cultivation parameters. The quantification was performed using a dot blot and the images analyzed using ImageQuant software (densitometry). The relative amount was then calculated based on a standard curve and normalized towards the process control (central point) to facilitate the evaluation of the effect of varying the factors considered. (*) process control, representing a set of parameters that, based on the literature and previous experiments, were known to lead to the production of RBD.). From the figure, reactor 1 from week 7 (7.1), gave the best relative percentage, around 25% in comparison with the process control. As a result of the good reproducibility, regardless of run week, batch variability or position in the Ambr 250 module, the results from the full experimental design can be attributed to changes in factor levels with confidence.
Phase 3: technology transfer & scale-up studies of the optimized production process
The optimized culture conditions obtained in the previous phase were then tech-transferred to PT Bio Farma, which had successfully scaled up the small-scale platform to a 5 L cultivation process (Figure 4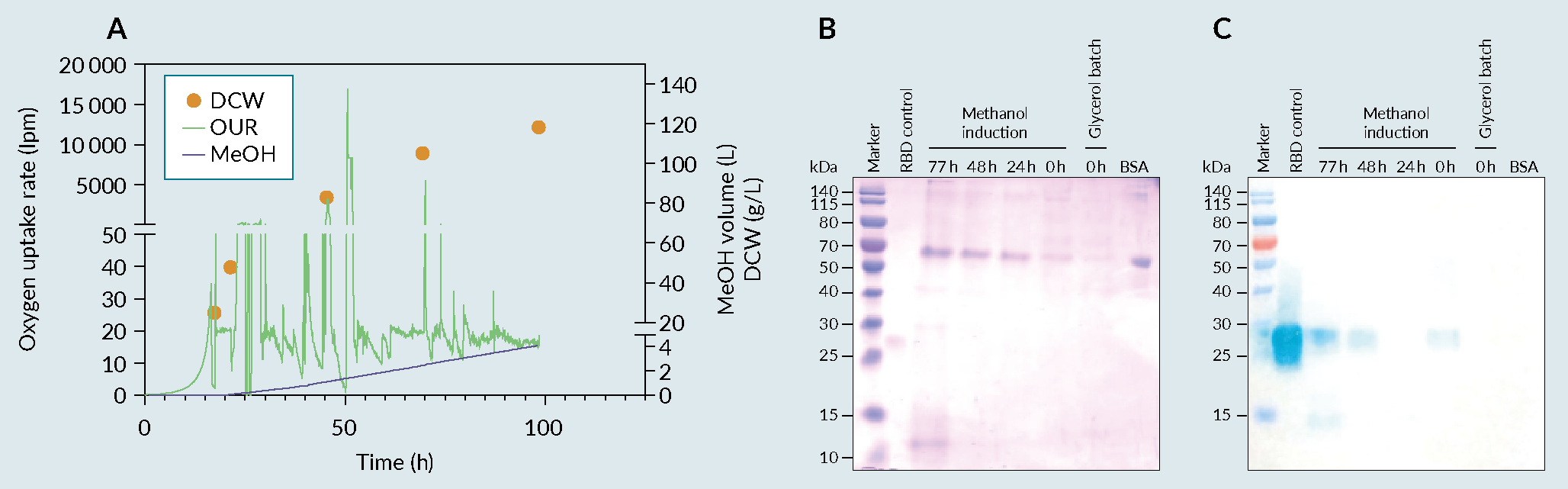 A) fermentation profile of cultivation in 5L fermenter, B) biochemical evaluation of produced receptor-binding domain (RBD) using an SDS-PAGE and C) a Western Blot (WB).). The secretion of the recombinant protein into the culture medium is essential for the easy recovery of proteins from a cell culture system. The produced RBD was then purified, using a PT Bio Farma in-house protocol, and its biochemical characteristics were evaluated (Figure 5
A) fermentation profile of cultivation in 5L fermenter, B) biochemical evaluation of produced receptor-binding domain (RBD) using an SDS-PAGE and C) a Western Blot (WB).). The secretion of the recombinant protein into the culture medium is essential for the easy recovery of proteins from a cell culture system. The produced RBD was then purified, using a PT Bio Farma in-house protocol, and its biochemical characteristics were evaluated (Figure 5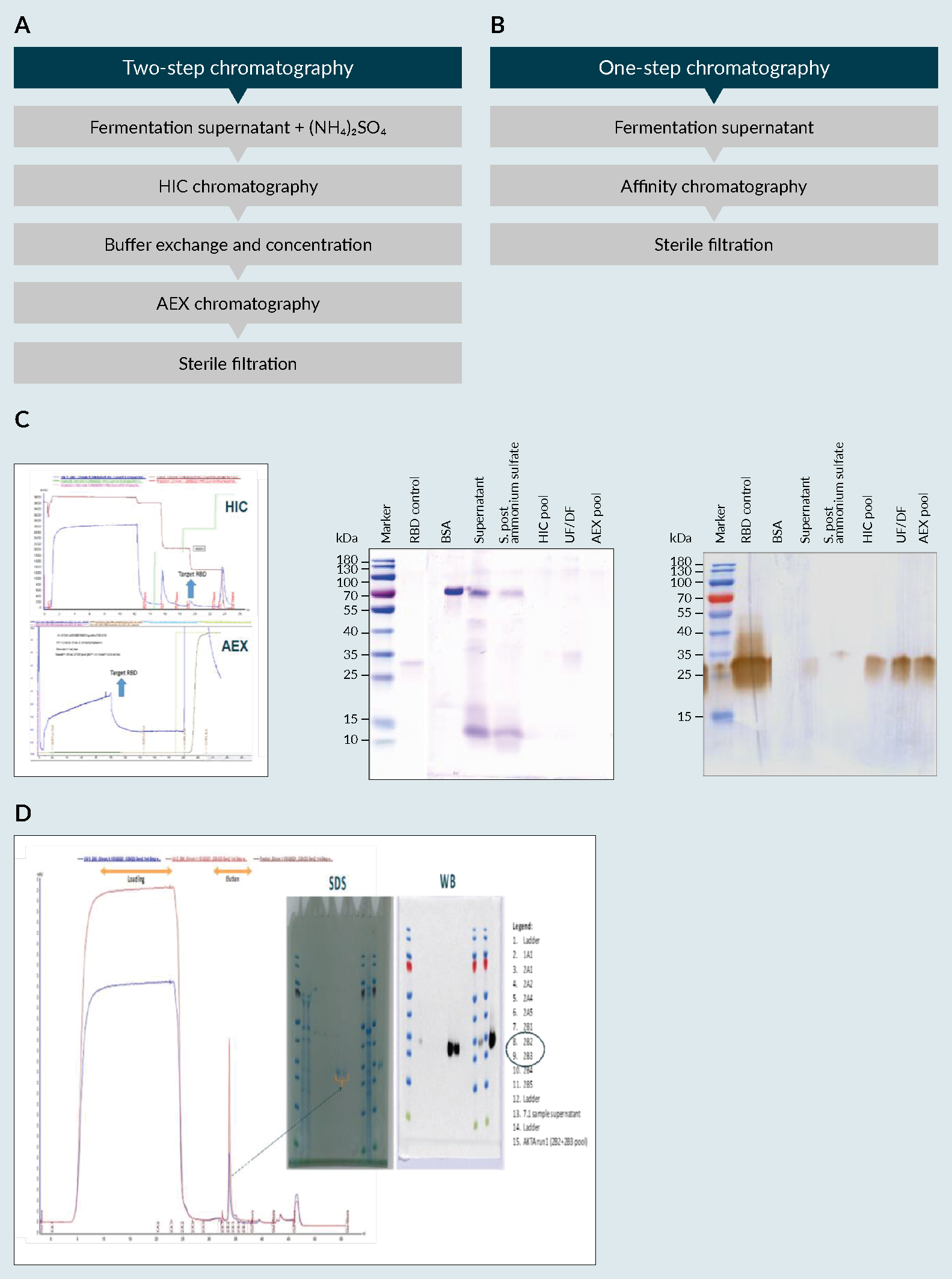 RBD purification processes using HIC/AEX and Affinity chromatography. A) flow Chart of the HIC/AEX purification process, B) flow Chart of the Affinity purification process C) HIC/AEX chromatograms and biochemical evaluation using SDS-PAGE and WB, RBD MW=25 to 32kDa D) affinity chromatogram and biochemical evaluation using SDS and WB. Ammonium Sulfate ((NH4)2SO4), Hydrophobic interaction chromatography (HIC), Anion exchange chromatography (AEX), Sodium dodecyl sulfate (SDS), Western Blot (WB), receptor-binding domain (RBD).A & 5C). The methodology consisted of two-step chromatography (hydrophilic interaction chromatography and anion-exchange). With a yield of approximately 350 mg/L of fermentation supernatant, the multi-step standard procedure used in the purification process allowed for an approximately 30% recovery of the target product (data not shown) with a purity more than 90% (data in accordance to [9]Lee J, Liu Z, Chen WH et al. Process Development and Scale-up Optimization of the SARS-CoV-2 Receptor Binding Domain–Based Vaccine Candidate, RBD219-N1C1. Appl. Microbiol. Biotechnol. 2021, 105, 4153–4165. [13]Chen WH, Pollet J, Strych U et al. Yeast-Expressed Recombinant SARS-CoV-2 Receptor Binding Domain RBD203-N1 as a COVID-19 Protein Vaccine Candidate. Protein Expr. Purif. 2022, 190, 106003.); however, the development of another robust methodology that could contribute to the readiness of the process due to its simplicity was deemed important to the success of the platform.
RBD purification processes using HIC/AEX and Affinity chromatography. A) flow Chart of the HIC/AEX purification process, B) flow Chart of the Affinity purification process C) HIC/AEX chromatograms and biochemical evaluation using SDS-PAGE and WB, RBD MW=25 to 32kDa D) affinity chromatogram and biochemical evaluation using SDS and WB. Ammonium Sulfate ((NH4)2SO4), Hydrophobic interaction chromatography (HIC), Anion exchange chromatography (AEX), Sodium dodecyl sulfate (SDS), Western Blot (WB), receptor-binding domain (RBD).A & 5C). The methodology consisted of two-step chromatography (hydrophilic interaction chromatography and anion-exchange). With a yield of approximately 350 mg/L of fermentation supernatant, the multi-step standard procedure used in the purification process allowed for an approximately 30% recovery of the target product (data not shown) with a purity more than 90% (data in accordance to [9]Lee J, Liu Z, Chen WH et al. Process Development and Scale-up Optimization of the SARS-CoV-2 Receptor Binding Domain–Based Vaccine Candidate, RBD219-N1C1. Appl. Microbiol. Biotechnol. 2021, 105, 4153–4165. [13]Chen WH, Pollet J, Strych U et al. Yeast-Expressed Recombinant SARS-CoV-2 Receptor Binding Domain RBD203-N1 as a COVID-19 Protein Vaccine Candidate. Protein Expr. Purif. 2022, 190, 106003.); however, the development of another robust methodology that could contribute to the readiness of the process due to its simplicity was deemed important to the success of the platform.
Towards a single-step chromatographic process with minimal sample treatment, we have developed an affinity chromatography method. Moreover, due to the selectivity of the resin used in affinity chromatography, the efficacy of the process was enhanced. Also, it is a simpler method, with the advantage of avoiding possible product losses along the purification steps. Given the selectivity of the resin, a sharp peak (280nm) was obtained (Figure 5B). At the same place, in red (260nm), a peak can be seen that is possibly related to DNA from dead cells and/or due to components on the supernatant that are also responsible for its colorization. Nonetheless, this didn’t affect the purity of the sample as seen from the biochemical analysis (Figure 5D), and can be easily removed using a polishing step, if necessary. With this simplified chromatographic method for product purification, we were able to achieve the same level of purity with similar yield, in a more time-efficient manner.
Phase 4: Animal studies for immunological efficiency evaluation of the vaccine
The immunogenic abilities of RBD were demonstrated in a mouse model. This immunogenicity study consisted of two stages. Firstly, five groups of seven mice were used to evaluate different amounts of RBD alone and in conjugation with an adjuvant (alum) in comparison with a positive control (CoronaVac [Sinovac]). The immunization scheme comprised two subcutaneous injections, with the second (boost) being administered after 21 days. The animal serum was collected at the end of 35 days and the antibody response was evaluated using commercial ELISA. The results showed that the immunological response is enhanced when the RBD is conjugated with the alum adjuvant. When combined with RBD, adjuvants with various characteristics elicit distinctive immunological profiles regarding the direction, duration, and strength of immune responses [14]Liang Z, Zhu H, Wang X et al. Adjuvants for Coronavirus Vaccines. Front. Immunol. 2020, 11.. However, IgG titers were not as high as expected, leading to a low % of antibody inhibition (data not shown). These results suggested further research on the antigen and adjuvant formulation is imperative to increase the efficacy of the vaccine. Therefore, a second attempt was performed (ten groups of seven mice each), where new antigen/adjuvant formulations were evaluated to accomplish higher titers and enhance the vaccine efficacy (Figure 6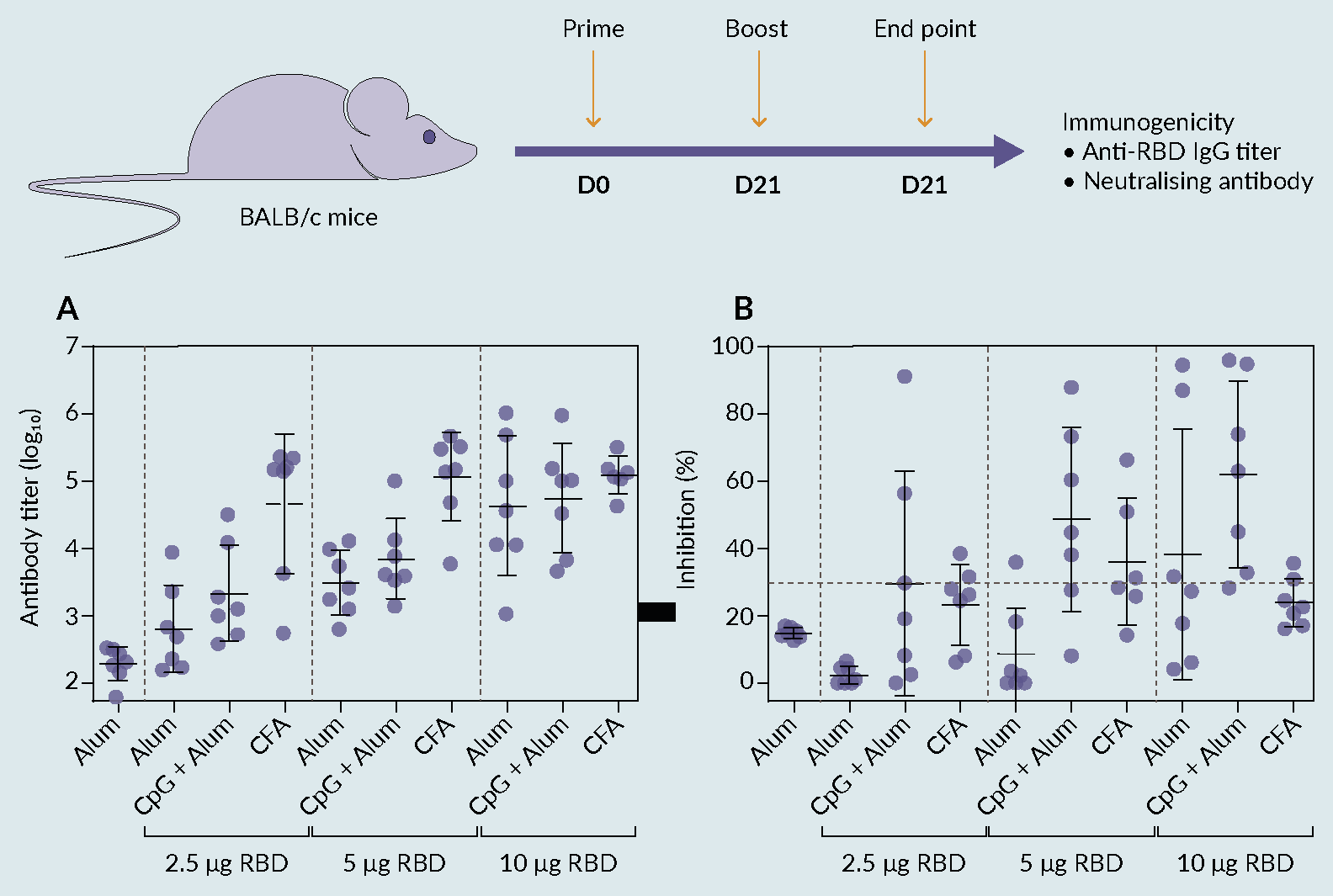 Figure 6. Immune responses induced by receptor binding domain (RBD) antigen. (A) groups of BALB/c mice (n = 7 per group) were vaccinated s.c. at the base of the tail on day 0 and again on day 21 with alum only or 2.5 μg, 5 μg, or 10 μg RBD formulated with Alum or Alum and CpG or CFA in a total volume of 200 μL/dose. Sera were obtained from blood taken on day 35. Antibody responses were then determined by ELISA and (B) antibody neutralization capacity was measured by SARS-CoV-2 Neutralization Antibody Detection Kit presented as percent inhibition. The antibody titers and percent inhibition of individual animals are presented with the mean value ± SD.). The selected adjuvants for this second attempt were: cytosine phosphoguanine (CpG) and AS03 (alhydrogel 1.3%), which have already been used in licensed vaccines [15]Shi S, Zhu H, Xia X, Liang Z, Ma X, Sun B. Vaccine Adjuvants: Understanding the Structure and Mechanism of Adjuvanticity. Vaccine 2019, 37, 3167–3178., and complete freund’s adjuvant for initial immunization [16]Grumstrup-Scott Guidelines for the Use of Adjuvants in Research Special Emphasis on Freund’s Adjuvant. 1989, 1–5.. From the obtained results, formulations with alum and CpG triggered a robust level of antigen-specific antibodies that possess neutralizing ability, which is in accordance with the results obtained by Chen et al. [13]Chen WH, Pollet J, Strych U et al. Yeast-Expressed Recombinant SARS-CoV-2 Receptor Binding Domain RBD203-N1 as a COVID-19 Protein Vaccine Candidate. Protein Expr. Purif. 2022, 190, 106003.. Also, it was observed that titers and antibody neutralization are dose- and adjuvant-dependent, giving promising insights for further formulation optimization.
Figure 6. Immune responses induced by receptor binding domain (RBD) antigen. (A) groups of BALB/c mice (n = 7 per group) were vaccinated s.c. at the base of the tail on day 0 and again on day 21 with alum only or 2.5 μg, 5 μg, or 10 μg RBD formulated with Alum or Alum and CpG or CFA in a total volume of 200 μL/dose. Sera were obtained from blood taken on day 35. Antibody responses were then determined by ELISA and (B) antibody neutralization capacity was measured by SARS-CoV-2 Neutralization Antibody Detection Kit presented as percent inhibition. The antibody titers and percent inhibition of individual animals are presented with the mean value ± SD.). The selected adjuvants for this second attempt were: cytosine phosphoguanine (CpG) and AS03 (alhydrogel 1.3%), which have already been used in licensed vaccines [15]Shi S, Zhu H, Xia X, Liang Z, Ma X, Sun B. Vaccine Adjuvants: Understanding the Structure and Mechanism of Adjuvanticity. Vaccine 2019, 37, 3167–3178., and complete freund’s adjuvant for initial immunization [16]Grumstrup-Scott Guidelines for the Use of Adjuvants in Research Special Emphasis on Freund’s Adjuvant. 1989, 1–5.. From the obtained results, formulations with alum and CpG triggered a robust level of antigen-specific antibodies that possess neutralizing ability, which is in accordance with the results obtained by Chen et al. [13]Chen WH, Pollet J, Strych U et al. Yeast-Expressed Recombinant SARS-CoV-2 Receptor Binding Domain RBD203-N1 as a COVID-19 Protein Vaccine Candidate. Protein Expr. Purif. 2022, 190, 106003.. Also, it was observed that titers and antibody neutralization are dose- and adjuvant-dependent, giving promising insights for further formulation optimization.
Conclusion
In this study, we report on the production of SARS CoV-2 spike protein RBD, using Komagataella phaffii as a hosting system. This yeast combines the speed and ease of highly efficient prokaryotic platforms with some key capabilities of mammalian systems, potentially reducing manufacturing costs and making it one of the most promising candidates for the expression of heterologous proteins in vaccine development [5]de Sá Magalhães S, Keshavarz‐Moore EP. Pastoris (Komagataella Phaffii) as a Cost‐effective Tool for Vaccine Production for Low‐ and Middle‐income Countries (Lmics). Bioeng. 2021, 8.. The fermentation yield of this vaccine prototype was 350 mg/L. The simplified purification process showed similar results to the in-house process. The purified RBD was of high purity (>90% by WB). Additionally, when formulated with alum along with CpG, it triggered a robust level of antigen-specific antibodies that possess neutralizing ability. Collectively, the data suggested this vaccine prototype is a suitable COVID-19 vaccine candidate antigen for technology transfer.
Discussion
In general, access to vaccine products and technologies is obtained through licensing agreements between technology/product donors to recipient manufacturers. However, in the context of the COVID-19 vaccine, it is challenging for LMIC to get access, not only to vaccine products to meet the country’s needs but also to the current know-how in vaccine manufacturing processes [18]de Bengy Puyvallée A, Storeng KT. COVAX, Vaccine Donations and the Politics of Global Vaccine Inequity. Global. Health 2022, 18, 1–14. [19]Paul Kagame, President, Rwanda; Emmanuel Macron, President, France; Cyril Ramaphosa, President, South Africa; Macky Sall, President, Senegal; Olaf Scholz, Chancellor, Germany; and Dr Tedros Adhanom Ghebreyesus, Director General, W. Sharing Technology and Supporting Innovation Is Not Just about Equity. It’s Also the Best Way to Stop Pandemics. (Accessed on 29 March 2023).. One of the potential factors could be that many technology owners have a high demand for collaboration at the same time and thus, the companies tend to choose their counterparts selectively for expansion of their vaccine production. One lesson learned from the current COVID-19 pandemic is that self–reliance in vaccine development and production in LMIC is an essential strategy to be embraced [20] [21]Hotez PJ, Bottazzi ME. Developing a Low-Cost and Accessible Covid-19 Vaccine for Global Health. PLoS Negl. Trop. Dis. 2020, 14, 1–6.. In this collaborative study, PT Bio Farma had the opportunity of working closely with experts in the field of vaccine bioprocess development at UCL. This has expedited the development of a microbial platform to produce COVID-19 vaccine candidates by addressing the knowledge gaps and establishing readiness against not only current but also future pandemic threats.
Translational Insight
Vaccination is critical for the prevention and control of infectious disease outbreaks. Being of paramount importance to global health, they are a key component of primary health care and an indisputable human right. Yet far too many people around the world have insufficient access to vaccines. The limited availability and affordability of vaccines to LMIC has created a need for solutions that will ensure effective, affordable vaccine production technology. With the potential for more pandemics, the urgency to expand the vaccine range has become even more evident. Transfer of know-how will therefore be critical to enable scale-up. This case study will act as a foundation for the establishment and development of a COVID-19 vaccine platform for industrial production.
Affiliations
Salome De Sa Magalhaes
Department of Biochemical Engineering,
Faculty of Engineering Sciences,
UCL, UK
Stephen A Morris
Department of Biochemical Engineering,
Faculty of Engineering Sciences,
UCL, UK
Shinta Kusumawardani
PT Bio Farma,
Bandung, Indonesia
Acep Wijayadikusumah Riza
PT Bio Farma,
Bandung, Indonesia
Neni Nurainy
PT Bio Farma,
Bandung, Indonesia
Eli Keshavarz-Moore
Department of Biochemical Engineering,
Faculty of Engineering Sciences,
UCL, UK
References
1. Choi EM. COVID-19 Vaccines for Low- And Middle-Income Countries. Trans. R. Soc. Trop. Med. Hyg. 2021, 115; 447–456. Crossref
2. Mihigo R, Okeibunor J, Cernuschi T, Petu A, Satoulou A, Zawaira F. Improving Access to Affordable Vaccines for Middle-Income Countries in the African Region. Vaccine 2019, 37, 2838–2842. Crossref
3. Engineering UD of B. The Future Vaccine Manufacturing Research Hub: Securing the Supply of Essential Vaccines. (Accessed on 29 March 2023). Crossref
4. Lan J, Ge J, Yu J et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. Crossref
5. de Sá Magalhães S, Keshavarz‐Moore EP. Pastoris (Komagataella Phaffii) as a Cost‐effective Tool for Vaccine Production for Low‐ and Middle‐income Countries (Lmics). Bioeng. 2021, 8. Crossref
6. Kriesi WPT. by Pichia Produced Products on the Market. (Accessed on 29 March 2023). Crossref
7. Invitrogen Corporation Pichia Fermentation Process Guidelines Overview Overview, Continued. Prog. Bot. 2002, 67, 1–11. Crossref
8. Kleanthous H, Silverman JM, Makar KW, Yoon IK, Jackson N, Vaughn DW. Scientific Rationale for Developing Potent RBD-Based Vaccines Targeting COVID-19. npj Vaccines 2021, 6, 1–10. Crossref
9. Lee J, Liu Z, Chen WH et al. Process Development and Scale-up Optimization of the SARS-CoV-2 Receptor Binding Domain–Based Vaccine Candidate, RBD219-N1C1. Appl. Microbiol. Biotechnol. 2021, 105, 4153–4165. Crossref
10. Dalvie NC, Biedermann AM, Rodriguez-Aponte SA et al. Scalable, Methanol-Free Manufacturing of the SARS-CoV-2 Receptor-Binding Domain in Engineered Komagataella Phaffii. Biotechnol. Bioeng. 2022, 119, 657–662. Crossref
11. Bowden GD, Pichler BJ, Maurer A. A Design of Experiments (DoE) Approach Accelerates the Optimization of Copper-Mediated 18F-Fluorination Reactions of Arylstannanes. Sci. Rep. 2019, 9, 1–10. Crossref
12. Peng X, Yang G, Shi Y, Zhou Y, Zhang M, Li S. Box–Behnken Design Based Statistical Modeling for the Extraction and Physicochemical Properties of Pectin from Sunflower Heads and the Comparison with Commercial Low-Methoxyl Pectin. Sci. Rep. 2020, 10, 1–10. Crossref
13. Chen WH, Pollet J, Strych U et al. Yeast-Expressed Recombinant SARS-CoV-2 Receptor Binding Domain RBD203-N1 as a COVID-19 Protein Vaccine Candidate. Protein Expr. Purif. 2022, 190, 106003. Crossref
14. Liang Z, Zhu H, Wang X et al. Adjuvants for Coronavirus Vaccines. Front. Immunol. 2020, 11. Crossref
15. Shi S, Zhu H, Xia X, Liang Z, Ma X, Sun B. Vaccine Adjuvants: Understanding the Structure and Mechanism of Adjuvanticity. Vaccine 2019, 37, 3167–3178. Crossref
16. Grumstrup-Scott Guidelines for the Use of Adjuvants in Research Special Emphasis on Freund’s Adjuvant. 1989, 1–5. Crossref
17. Kohler J, Wong A, Tailor L. Improving Access to COVID-19 Vaccines: An Analysis of TRIPS Waiver Discourse among WTO Members, Civil Society Organizations, and Pharmaceutical Industry Stakeholders. Health Hum. Rights 2022, 24, 159–175. Crossref
18. de Bengy Puyvallée A, Storeng KT. COVAX, Vaccine Donations and the Politics of Global Vaccine Inequity. Global. Health 2022, 18, 1–14. Crossref
19. Paul Kagame, President, Rwanda; Emmanuel Macron, President, France; Cyril Ramaphosa, President, South Africa; Macky Sall, President, Senegal; Olaf Scholz, Chancellor, Germany; and Dr Tedros Adhanom Ghebreyesus, Director General, W. Sharing Technology and Supporting Innovation Is Not Just about Equity. It’s Also the Best Way to Stop Pandemics. (Accessed on 29 March 2023). Crossref
20. Olufadewa II, Adesina MA, Ekpo MD et al. Lessons from the Coronavirus Disease 2019 (COVID-19) Pandemic Response in China, Italy, and the U.S.: A Guide for Africa and Low- and Middle-Income Countries. Glob. Heal. J. 2021, 5, 56–61. Crossref
21. Hotez PJ, Bottazzi ME. Developing a Low-Cost and Accessible Covid-19 Vaccine for Global Health. PLoS Negl. Trop. Dis. 2020, 14, 1–6. Crossref
Authorship & Conflict of Interest
Contributions: All named authors take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Acknowledgements: None.
Disclosure and potential conflicts of interest:The authors have no conflicts of interest.
Funding declaration: The authors received financial support for the research, authorship and/or publication of this article from Engineering and Physical Sciences Research Council (EPSRC), Department of Health and Social Care, Future Vaccine Research Manufacturing Hub (Vax-Hub, EP/R013756/1)] for grants, from PT Bio Farma for provision of cell lines and UCL for written tech transfer
Article & copyright information
Copyright: Published by Vaccine Insights under Creative Commons License Deed CC BY NC ND 4.0 which allows anyone to copy, distribute, and transmit the article provided it is properly attributed in the manner specified below. No commercial use without permission.
Attribution: Copyright © 2023 Keshavarz-Moore E. Published by Vaccine Insights under Creative Commons License Deed CC BY NC ND 4.0.
Article source: Invited; externally peer reviewed.
Submitted for peer review:Apr 3 2023;Revised manuscript received:May 10 2023; Publication date: Jun 8 2023.