Chromatographic purification with CIMmultus™ Oligo dT increases mRNA stability
Cell & Gene Therapy Insights 2021; 7(9), 1207–1216
10.18609/cgti.2021.161
One of the major challenges of mRNA based vaccines has been their requirement for distribution and storage at extremely low temperatures, indicating that exposure of mRNA to suboptimal physico-chemical conditions can result in degradation and loss of potency; it is unclear whether this is due to instability of mRNA drug substance, or LNP-encapsulated mRNA, or both. In this study we compare the stability of model mRNA drug substance (eGFP, 995 nt) prepared by affinity chromatography with the stability of mRNA purified by precipitation. We show that both purification methods lead to highly pure mRNA drug substance, however, mRNA purified by chromatography remains stable for 28 days at 37°C, whereas mRNA purified by precipitation is subject to significant degradation under the same storage conditions. We conclude that chromatography eliminates elements and/or conditions with adverse impact on the quality of mRNA to a greater extent than precipitation method and that choosing appropriate purification strategy is crucial not only to achieve target purity but also to obtain a stable product with retained integrity.
Strengths of mRNA technology were recently demonstrated by the extraordinarily rapid development and clinical success of two vaccines against SARS-CoV-2 and variants [1]Baden LR, El Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384(5):403-416., [2]Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385(7):585-594., [3]Verbeke R, Lentacker I, De Smedt SC, Dewitte H. The dawn of mRNA vaccines: The COVID-19 case. J Control Release. 2021;333:511-520.. However, the challenges of global roll-out exposed limitations associated with (ultra)cold-storage requirements [4]Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J Pharm Sci. 2021;110(3):997-1001Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J Pharm Sci. 2021;110(3):997-1001. Moderna’s Spikevax vaccine requires long-term storage between -15 and -25°C and BioNTech/Pfizer’s Comirnaty vaccine between -60 and -90°C [5]Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.. Both mRNA vaccines use lipid nanoparticle (LNP) formulation of mRNA drug substance and it is unclear what factors lead to the different temperature requirements for long-term storage of the two vaccines - whether these arise from instability of mRNA drug substance, LNP-encapsulated mRNA (‘drug product’), or a combination thereof [5]Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.. A recent publication evaluated stability of LNP-encapsulated mRNA and demonstrated its stability at 25°C for 7 days, suggesting that encapsulation itself has a stabilizing effect on mRNA [6]Zhang NN, Li XF, Deng YQ, et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182(5):1271-1283.e1Zhang NN, Li XF, Deng YQ, et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182(5):1271-1283.e1; instability of mRNA vaccines could thus potentially stem from instability of mRNA drug substance itself.
With the proviso that mRNA stability can be highly sequence-dependent [7]Leppek K, Byeon GW, Kladwang W, et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Preprint. bioRxiv. 2021;2021.03.29.43758 , [8]Wayment-Steele HK, Kim DS, Choe CA, et al. Theoretical basis for stabilizing messenger RNA through secondary structure design. Preprint. bioRxiv. 2021;2020.022.262931. Published 2021 Feb 19, it is possible that for a given sequence, impurity profile and/or purification approach, which could induce physico-chemical or mechanical stress, lead to instability of mRNA drug substance. Ribonucleic acids (RNA) are relatively unstable biomolecules, especially when compared to deoxyribonucleic acids (DNA). Degradation rates are very heterogeneous in solution because of the variety of local RNA conformations, which determine hydrolysis rates [9]AbouHaidar MG, Ivanov IG. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z Naturforsch C J Biosci. 1999;54(7-8):542-548.AbouHaidar MG, Ivanov IG. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z Naturforsch C J Biosci. 1999;54(7-8):542-548.. Hydrolytic cleavage of the phosphodiester bond is catalyzed by some metallic complexes, including with Mg2+, one of the critical cofactors required during production of mRNA [9]AbouHaidar MG, Ivanov IG. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z Naturforsch C J Biosci. 1999;54(7-8):542-548.AbouHaidar MG, Ivanov IG. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z Naturforsch C J Biosci. 1999;54(7-8):542-548., some reports even proposed that full protection from atmosphere is needed to prevent hydrolysis for long term storage [10]Fabre AL, Colotte M, Luis A, Tuffet S, Bonnet J. An efficient method for long-term room temperature storage of RNA. Eur J Hum Genet. 2014;22(3):379-385.. mRNA stability is therefore highly dependent on the chemical environment to which it is exposed during production process.
An IVT reaction mixture contains a number of components, including enzymes, residual NTPs and DNA template, as well as small molecule additives used to improve mRNA yield (spermidine, Triton, etc.), that need to be removed during the manufacturing process. Traditional lab-scale purification methods are based on DNA removal by DNAse digestion followed by lithium chloride (LiCl) or ethanol precipitation, but these methods are chemically harsh, introduce toxic chemicals into the final product and are challenging to scale-up [11]Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200..
Chromatography offers a highly selective as well as scalable approach to purification, which when fully optimized delivers high purity of therapeutic agents. Due to its large size (1000 nt mRNA is approximately 400 kDa, nearly 3-times larger than an IgG), mRNA diffusion coefficient is low, rendering traditional chromatographic approaches, which depend on diffusive mass transport, less suitable as a purification tool for this therapeutic class [12]Gagnon P. Purification of Nucleic Acids A Handbook for Purification of DNA Plasmids and mRNA for Gene Therapy and Vaccines.; 2020.Gagnon P. Purification of Nucleic Acids A Handbook for Purification of DNA Plasmids and mRNA for Gene Therapy and Vaccines.; 2020.. Convective-flow purification media, such as monoliths, are more suitable for purification of such large biomolecules, providing higher binding capacity, faster purification cycles and lower shear [11]Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200., [13]Kramberger P, Urbas L, Štrancar A. Downstream processing and chromatography based analytical methods for production of vaccines, gene therapy vectors, and bacteriophages. Hum Vaccin Immunother. 2015;11(4):1010-1021. doi:10.1080/21645515.2015.1009817Kramberger P, Urbas L, Štrancar A. Downstream processing and chromatography based analytical methods for production of vaccines, gene therapy vectors, and bacteriophages. Hum Vaccin Immunother. 2015;11(4):1010-1021. doi:10.1080/21645515.2015.1009817, [14]Yamamoto S, Yoshimoto N, Nishizumi Y. Theoretical background of monolithic short layer ion-exchange chromatography for separation of charged large biomolecules or bioparticles. J Chromatogr A. 2009;1216(13):2612-2615.Yamamoto S, Yoshimoto N, Nishizumi Y. Theoretical background of monolithic short layer ion-exchange chromatography for separation of charged large biomolecules or bioparticles. J Chromatogr A. 2009;1216(13):2612-2615., [15]Podgornik A, Yamamoto S, Peterka M, Krajnc NL. Fast separation of large biomolecules using short monolithic columns. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;927:80-89. doi:10.1016/j.jchromb.2013.02.004Podgornik A, Yamamoto S, Peterka M, Krajnc NL. Fast separation of large biomolecules using short monolithic columns. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;927:80-89. doi:10.1016/j.jchromb.2013.02.004, [16]Yamamoto S, Nakamura M, Tarmann C, Jungbauer A. Retention studies of DNA on anion-exchange monolith chromatography Binding site and elution behavior. J Chromatogr A. 2007;1144(1):155-160.Yamamoto S, Nakamura M, Tarmann C, Jungbauer A. Retention studies of DNA on anion-exchange monolith chromatography Binding site and elution behavior. J Chromatogr A. 2007;1144(1):155-160.. For mRNA constructs containing a polyadenylic acid (PolyA) tail, the mRNA can be isolated from the IVT mixture in a pseudo-affinity mode, using affinity of chromatography support-immobilized poly-deoxythymidine for mRNA containing PolyA tail. For constructs that do not contain a polyA tail, charge or hydrogen bonding interactions can be employed [12]Gagnon P. Purification of Nucleic Acids A Handbook for Purification of DNA Plasmids and mRNA for Gene Therapy and Vaccines.; 2020.Gagnon P. Purification of Nucleic Acids A Handbook for Purification of DNA Plasmids and mRNA for Gene Therapy and Vaccines.; 2020..
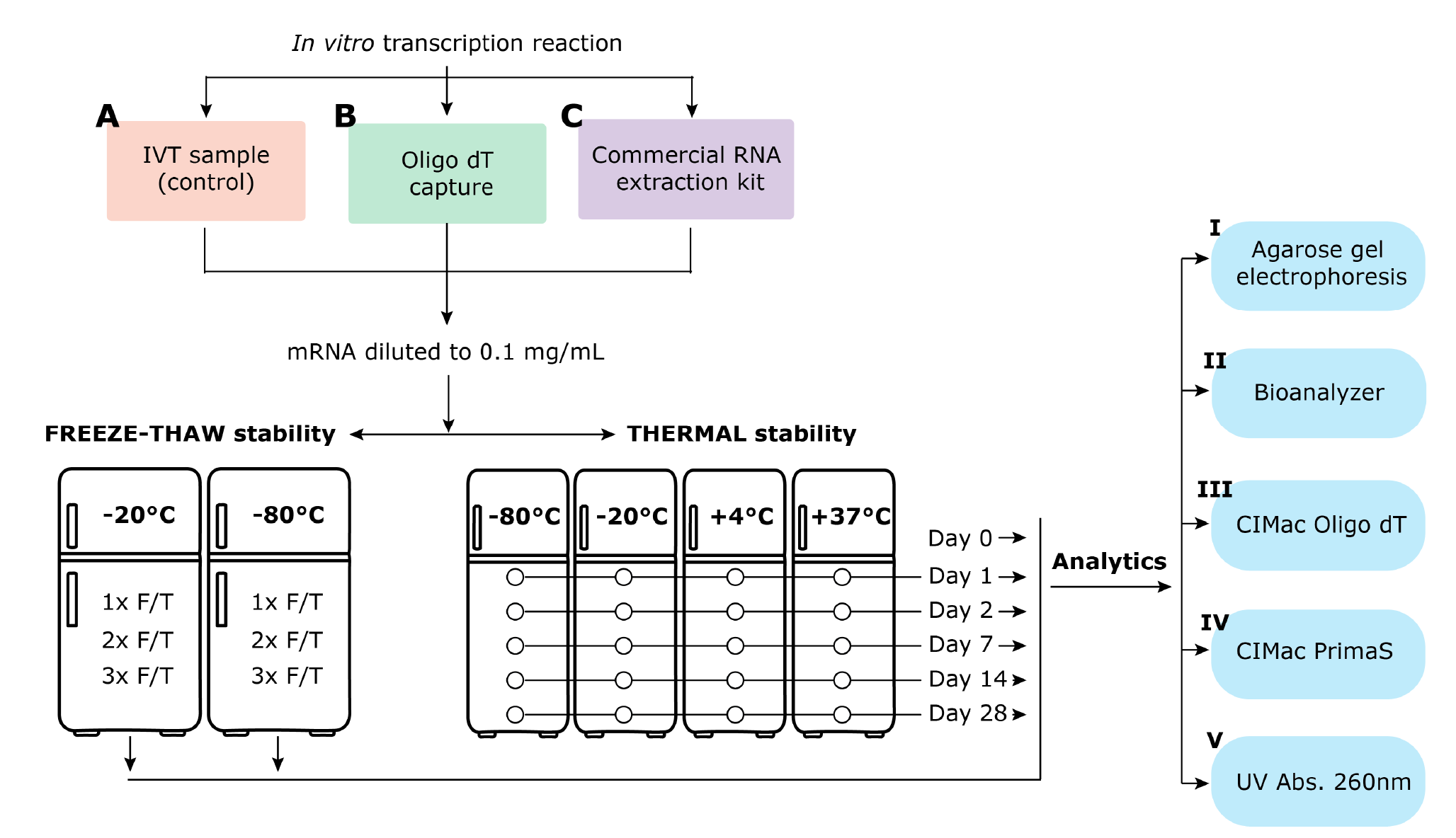
In this study we evaluated the contribution of purification methodology to quality of mRNA drug substance, using stability as an index. Two purification approaches were evaluated, both expected to yield highly pure mRNA: RNA extraction kit was chosen as a standard RNA purification tool used in many laboratories working with RNA, and monolithic chromatography media was selected for their ability to support rapid high-resolution separation of very large molecules in a low-shear environment [11]Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200., [13]Kramberger P, Urbas L, Štrancar A. Downstream processing and chromatography based analytical methods for production of vaccines, gene therapy vectors, and bacteriophages. Hum Vaccin Immunother. 2015;11(4):1010-1021. doi:10.1080/21645515.2015.1009817Kramberger P, Urbas L, Štrancar A. Downstream processing and chromatography based analytical methods for production of vaccines, gene therapy vectors, and bacteriophages. Hum Vaccin Immunother. 2015;11(4):1010-1021. doi:10.1080/21645515.2015.1009817, [14]Yamamoto S, Yoshimoto N, Nishizumi Y. Theoretical background of monolithic short layer ion-exchange chromatography for separation of charged large biomolecules or bioparticles. J Chromatogr A. 2009;1216(13):2612-2615.Yamamoto S, Yoshimoto N, Nishizumi Y. Theoretical background of monolithic short layer ion-exchange chromatography for separation of charged large biomolecules or bioparticles. J Chromatogr A. 2009;1216(13):2612-2615., [15]Podgornik A, Yamamoto S, Peterka M, Krajnc NL. Fast separation of large biomolecules using short monolithic columns. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;927:80-89. doi:10.1016/j.jchromb.2013.02.004Podgornik A, Yamamoto S, Peterka M, Krajnc NL. Fast separation of large biomolecules using short monolithic columns. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;927:80-89. doi:10.1016/j.jchromb.2013.02.004, [16]Yamamoto S, Nakamura M, Tarmann C, Jungbauer A. Retention studies of DNA on anion-exchange monolith chromatography Binding site and elution behavior. J Chromatogr A. 2007;1144(1):155-160.Yamamoto S, Nakamura M, Tarmann C, Jungbauer A. Retention studies of DNA on anion-exchange monolith chromatography Binding site and elution behavior. J Chromatogr A. 2007;1144(1):155-160.. Stability of mRNA at 37°C (optimal temperature for activity of RNAses), 4°C (typical refrigeration temperature), -20°C and -80°C was assessed by a selection of standard physicochemical analytical techniques used for mRNA drug substance quality attribute assessment (UV spectroscopy (A260 nm) for content determination, agarose gel electrophoresis for molecular mass and RNA integrity assessment, chip capillary electrophoresis for quantification of fragmentation) [4]Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J Pharm Sci. 2021;110(3):997-1001Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J Pharm Sci. 2021;110(3):997-1001, [5]Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586., [6]Zhang NN, Li XF, Deng YQ, et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182(5):1271-1283.e1Zhang NN, Li XF, Deng YQ, et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182(5):1271-1283.e1, [17]Poveda C, Biter AB, Bottazzi ME, Strych U. Establishing Preferred Product Characterization for the Evaluation of RNA Vaccine Antigens. Vaccines (Basel). 2019;7(4):131. Published 2019 Sep 27., and two HPLC methods which are reported for the first time: quantification of poly-adenylated mRNA using CIMac™ Oligo dT and purity assessment of mRNA by CIMac PrimaS™. Freeze-thaw stability was also assessed after each purification approach using the same set of analytical methods. For the full materials and methods, please refer to the supplementary data for this article.
Results & discussion
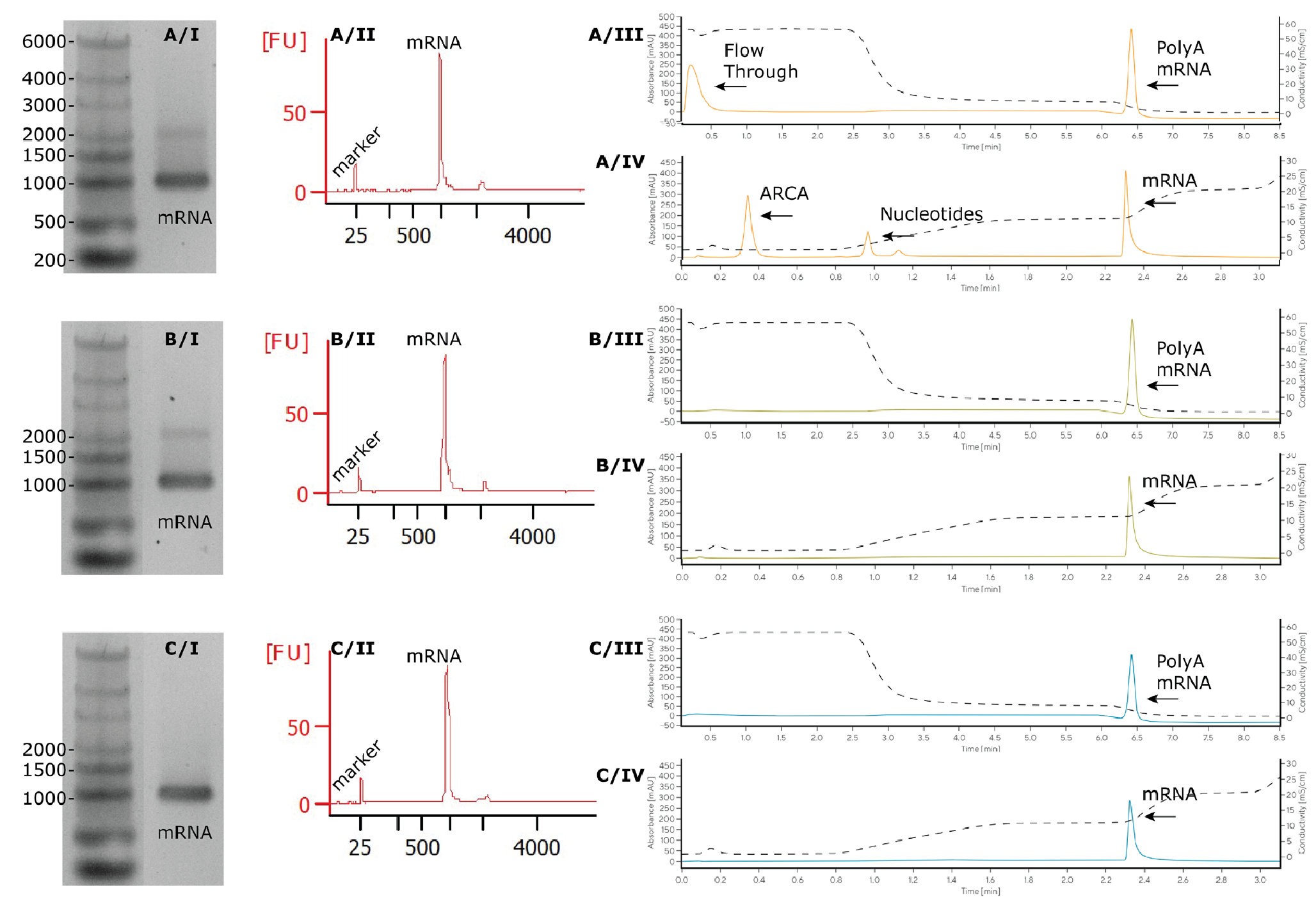
After IVT production of mRNA from a plasmid encoding for eGFP (950 nucleotides) and poly-adenosine tail (45 nucleotides) according to a standard IVT protocol (Figure 1
mRNA at expected molecular size (995 nt) was observed by AGE and BioAnalyzer for all samples. Oligo dT eluate showed a minor band at 2000 nt by both AGE and BioAnalyzer, corresponding to a dimeric form of mRNA, which disappeared with denaturing the sample by heating it at 70°C for 5 min (Supplementary Figure 1). For diluted IVT, analytical affinity chromatography (CIMac™ Oligo dT) revealed the expected flow-through peak, corresponding to UV-absorbing IVT reaction components (nucleotides, enzymes), and the elution peak, corresponding to polyadenylated RNA. Peak area corresponded to expected mRNA concentration (0.1 mg/mL). mRNA purified by affinity chromatography and extraction kit only showed elution peak at expected concentration and no flow-through peak. Similarly, CIMac PrimaS™ chromatogram demonstrated the presence of nucleotide-like reaction components (capping reagent and residual NTPs) and RNA in the IVT sample, but only RNA in samples purified by affinity chromatography and extraction kit (Figure 2
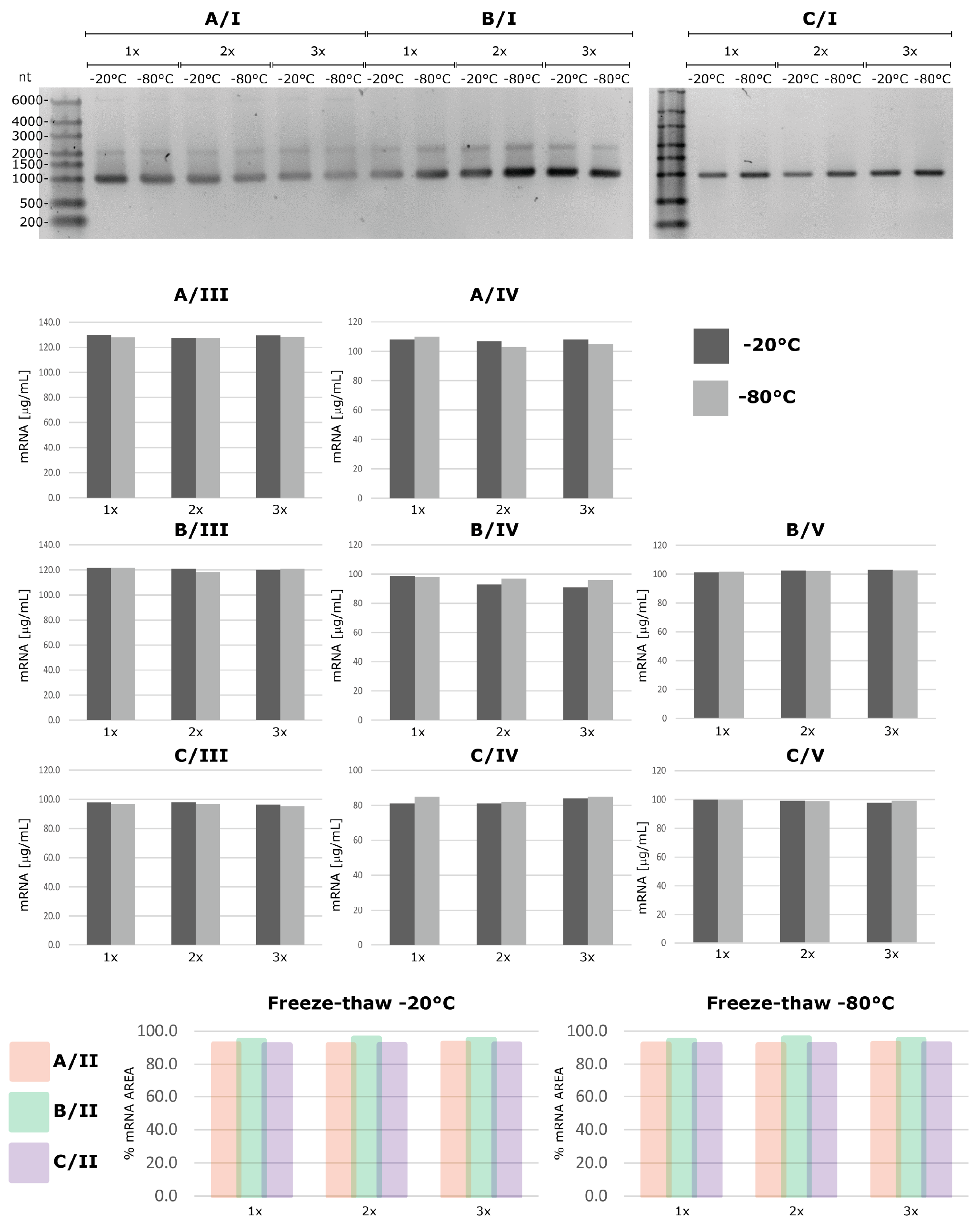
Stability of mRNA prepared according to the three methods was then evaluated at different temperatures over a period of 28 days, and freeze-thaw stability at -20°C/-80 was evaluated for up to three freeze-thaw cycles (Figure 1).
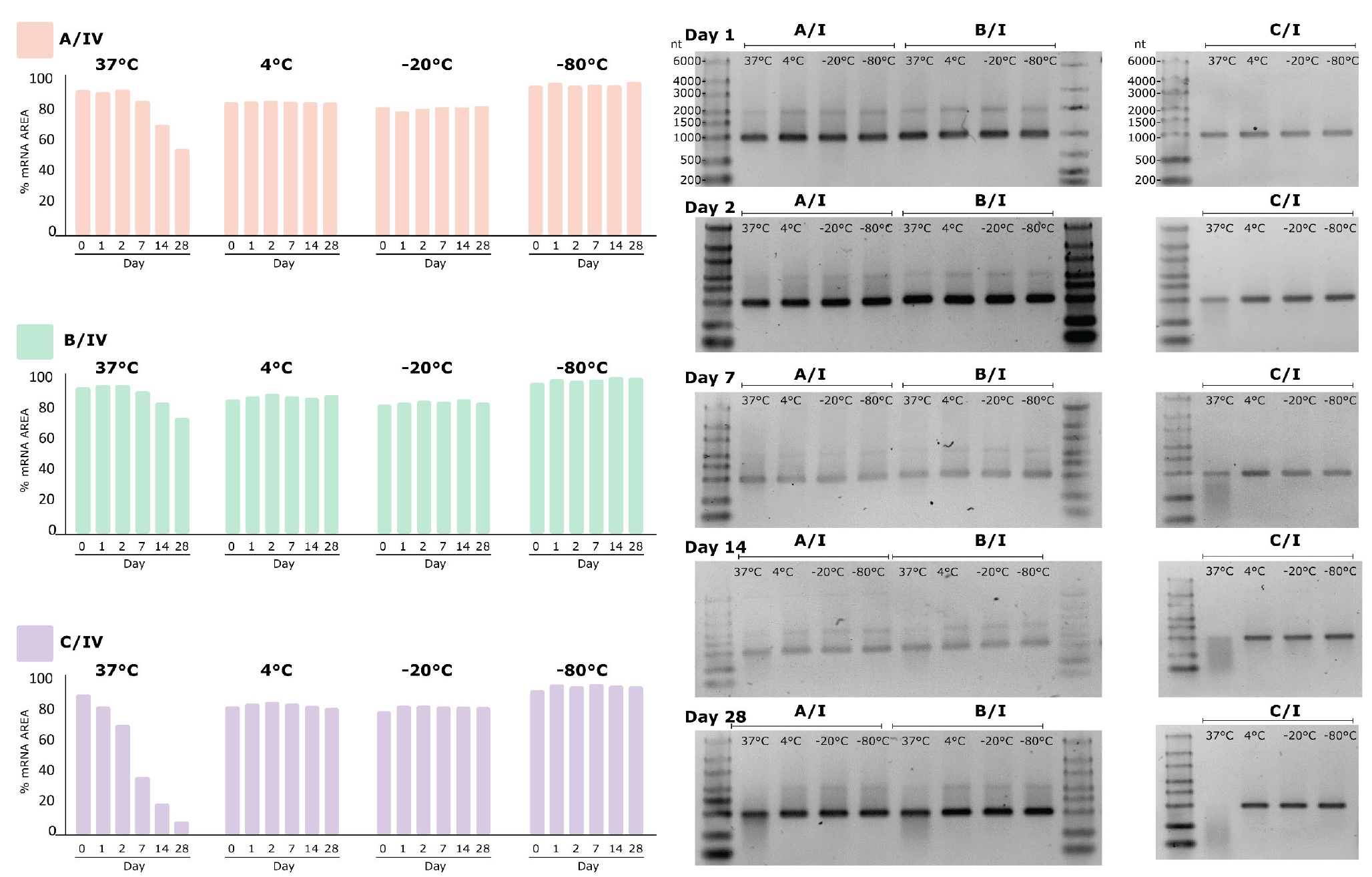
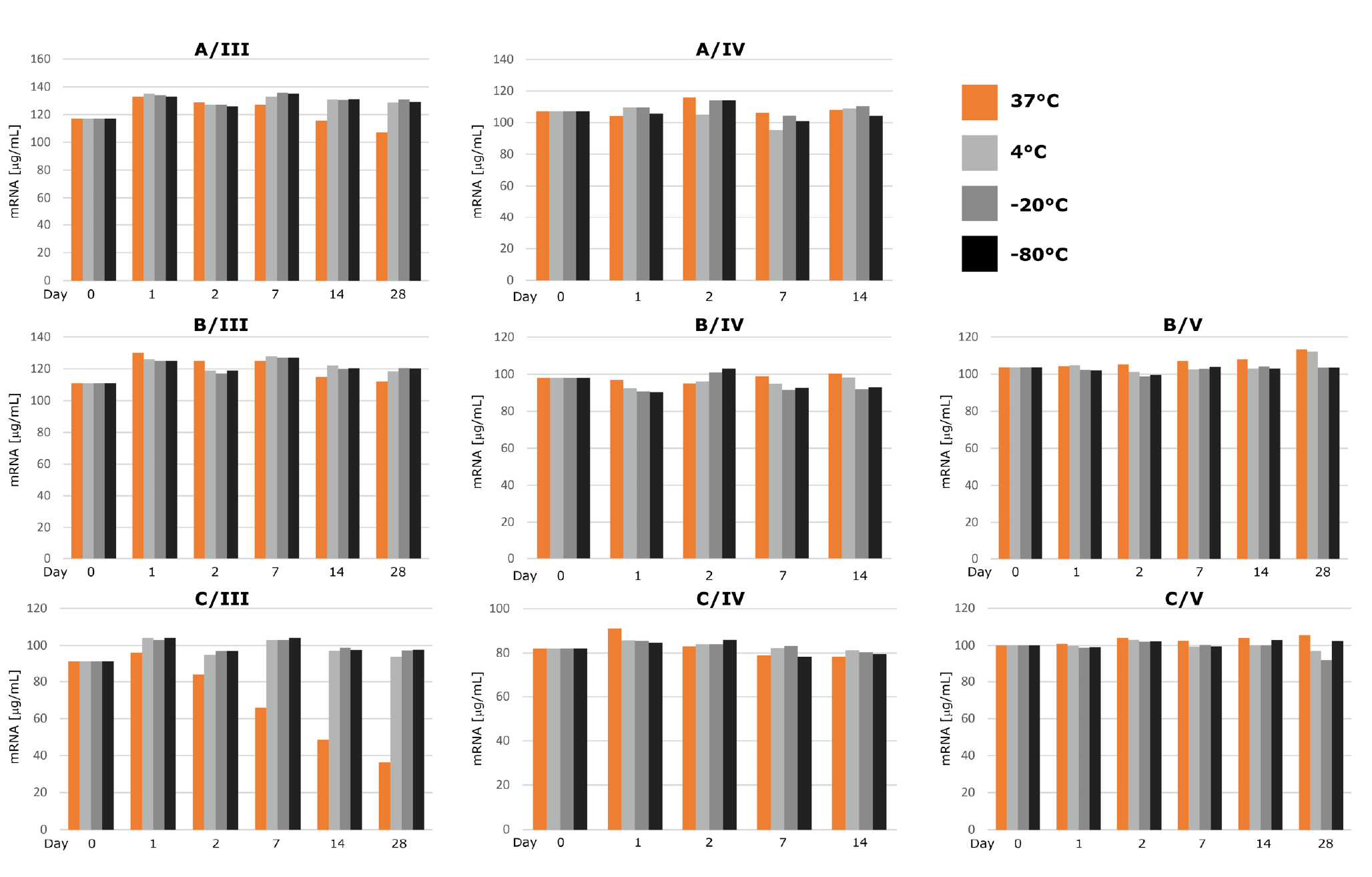
All three preparation methods resulted in mRNA that was stable at -80, -20 and 4°C for up to 28 days, and up to three freeze-thaw cycles at -20°C and -80°C (Figure 3


Chromatographic analysis by CIMac™ Oligo dT (labeled with roman number III in all figures) was in close agreement with electrophoretic results. The content of mRNA prepared by all three methods did not differ significantly for incubation temperatures of -80/-20/4°C, but a significant decrease in content (from starting 90 µg/ml to 38 to µg/ml by day 28) was observed for mRNA purified by extraction kit, but not other methods, when incubated at 37°C for up to 28 days (Figure 5 C/III). CIMac PrimaS™ analysis, which resolves mRNA from NTPs, capping reagent and plasmid, resulted in mRNA peak intensity which did not change with incubation time or temperature (Figure 5C/IV). Interestingly, analysis of peak corresponding to residual ARCA capping reagent (retention time of 0.4 min, Figure S3) in diluted IVT sample indicated a shift to 0.8 min observed at 37°C from day 1 onwards, suggesting possible degradation of the reagent. Although IVT reactions are seldomly carried out beyond 3-6 hours, stability of reactants under IVT conditions (e.g. 37°C) should nonetheless be evaluated to avoid potential safety issues due to incorporation of degradation products into nascent mRNA.
As expected, UV absorbance of stability samples resulted in no apparent difference in content as measured by absorbance at 260 nm even when other analytical methods showed severe degradation (Figure 5 (B/V and C/V)). Content by UV is one of the critical quality attributes assessed for release of mRNA vaccines [5]Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586.; our results point to the need to evaluate the UV results in conjunction with other purity methods, as it is not indicative of content of intact target molecule.
Conclusions
In this study, we compared stability of mRNA drug substance (995 nt) prepared by chromatographic method using CIMmultus Oligo dT column versus a precipitation technique using commercial RNA extraction kit. mRNA purified by chromatography was shown to remain stable for 28 days at 37°C, whereas purification by precipitation led to significant degradation of mRNA at 37°C, detectable by electrophoretic (AGE and BioAnalyzer), chromatographic (CIMac™ Oligo dT), but not UV-spectrophotometric method. Although follow-up studies on a wider range of mRNA sizes and modalities are warranted in the future, our results demonstrate the need for careful selection of purification strategy during development of mRNA therapeutics, which should be based on consideration of long-term stability impact, as well as scalability and compatibility with GMP standards.
References
1. Baden LR, El Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384(5):403-416. Crossref
2. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385(7):585-594. Crossref
3. Verbeke R, Lentacker I, De Smedt SC, Dewitte H. The dawn of mRNA vaccines: The COVID-19 case. J Control Release. 2021;333:511-520. Crossref
4. Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J Pharm Sci. 2021;110(3):997-1001 Crossref
5. Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586. Crossref
6. Zhang NN, Li XF, Deng YQ, et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182(5):1271-1283.e16. Crossref
7. Leppek K, Byeon GW, Kladwang W, et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Preprint. bioRxiv. 2021;2021.03.29.437587. Crossref
8. Wayment-Steele HK, Kim DS, Choe CA, et al. Theoretical basis for stabilizing messenger RNA through secondary structure design. Preprint. bioRxiv. 2021;2020.08.22.262931. Published 2021 Feb 19 Crossref
9. AbouHaidar MG, Ivanov IG. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z Naturforsch C J Biosci. 1999;54(7-8):542-548. Crossref
10. Fabre AL, Colotte M, Luis A, Tuffet S, Bonnet J. An efficient method for long-term room temperature storage of RNA. Eur J Hum Genet. 2014;22(3):379-385. Crossref
11. Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine. 2021;39(16):2190-2200. Crossref
12. Gagnon P. Purification of Nucleic Acids A Handbook for Purification of DNA Plasmids and mRNA for Gene Therapy and Vaccines.; 2020. Crossref
13. Kramberger P, Urbas L, Štrancar A. Downstream processing and chromatography based analytical methods for production of vaccines, gene therapy vectors, and bacteriophages. Hum Vaccin Immunother. 2015;11(4):1010-1021. doi:10.1080/21645515.2015.1009817 Crossref
14. Yamamoto S, Yoshimoto N, Nishizumi Y. Theoretical background of monolithic short layer ion-exchange chromatography for separation of charged large biomolecules or bioparticles. J Chromatogr A. 2009;1216(13):2612-2615. Crossref
15. Podgornik A, Yamamoto S, Peterka M, Krajnc NL. Fast separation of large biomolecules using short monolithic columns. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;927:80-89. doi:10.1016/j.jchromb.2013.02.004 Crossref
16. Yamamoto S, Nakamura M, Tarmann C, Jungbauer A. Retention studies of DNA on anion-exchange monolith chromatography Binding site and elution behavior. J Chromatogr A. 2007;1144(1):155-160. Crossref
17. Poveda C, Biter AB, Bottazzi ME, Strych U. Establishing Preferred Product Characterization for the Evaluation of RNA Vaccine Antigens. Vaccines (Basel). 2019;7(4):131. Published 2019 Sep 27. Crossref
18. Černigoj U, Štrancar A. Scale-Up of Plasmid DNA Downstream Process Based on Chromatographic Monoliths. Methods Mol Biol. 2021;2197:167-192. Crossref
Affiliations
Matevž Korenč
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Nina Mencin
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Jasmina Puc
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Janja Skok
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Kristina Šprinzar Nemec
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Anže Martinčič Celjar
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Pete Gagnon
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Aleš Štrancar
BIA Separations d.o.o., a Sartorius company, Mirce 21, 5270 Ajdovščina, Slovenia
Rok Sekirnik
Author for Correspondence:
Rok Sekirnik, BIA Separations d.o.o., Mirce 21, 5270 Ajdovščina, Slovenia
Tel.: +386 59 699 500;
Fax: +386 59 699 599
E-mail: rok.sekirnik@biaseparations.com
Authorship & Conflict of Interest
Contributions: All named authors take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Acknowledgements: None.
Disclosure and potential conflicts of interest: All authors are employees of BIA Separations d.o.o., a Sartorius company. The authors declare that they have no other conflicts of interest.
Funding declaration: BioMay AG provided study materials (cell paste containing eGFP plasmid). The authors received no other financial support for the research, authorship and/or publication of this article.
Article & copyright information
Copyright: Published by Cell and Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0 which allows anyone to copy, distribute, and transmit the article provided it is properly attributed in the manner specified below. No commercial use without permission.
Attribution: Copyright © 2021 BIA Separations d.o.o. Published by Cell and Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0.
Article source: Invited, peer reviewed.
Submitted for peer review: Aug 23 2021; Revised manuscript received: Oct 1 2021; Publication date: 13 Oct 2021.