Quantitative analysis of lipids and nucleic acids in lipid nanoparticles using monolithic column
Cell & Gene Therapy Insights 2024; 10(6), 867–878
DOI: 10.18609/cgti.2024.097
Lipid nanoparticles, a promising platform for drug delivery, effectively encapsulating and protecting nucleic acids. Comprising cationic or ionizable lipids, helper lipids, and PEGylated lipids, LNPs facilitate cellular uptake and control of payload release. The composition of lipids is crucial for optimizing LNP formulations, necessitating robust analytical methods. This study presents the development of a reverse phase liquid chromatography method to separate and quantify lipids and nucleic acid in LNP formulations. The method utilizes a PATfix® analytical system equipped with an evaporative light scattering detector and a monolith CIMac™ C4 HLD chromatographic column. The method demonstrated efficient lipid separation and detection, with validation following international chromatography handbook guidelines, highlighting its sensitivity, linearity, precision, and accuracy. Furthermore, the method’s suitability for quantitative analysis was verified by assessing lipid ratios in various LNP formulations, confirming its applicability for monitoring lipid composition throughout the LNPs’ manufacturing process.
Lipid nanoparticles (LNPs) have emerged as versatile drug delivery systems capable of encapsulating various payloads, including different nucleic acids [1,2]. LNPs offer numerous advantages, such as protection of the payload from degradation, enhanced cellular uptake, and controlled release, making them promising candidates for therapeutic applications [3].
Lipid nanoparticles (LNPs) typically comprise cationic or ionizable lipids, helper lipids, and PEGylated lipids [4–6]. Cationic or ionizable lipids play a crucial role in enhancing the binding and transfection efficiency of RNA. Cationic lipids, such as DOTAP, possess a permanently charged headgroup, while ionizable lipids, such as SM-102, acquire charge in lower pH environments (around a pH of 6–7) and remain uncharged at physiological pH levels (around 7.4). The positively charged headgroups of these lipids interact electrostatically with the negatively charged phosphate backbone of RNA, facilitating better RNA-LNP association. The manufacturing of LNPs typically occurs under acidic conditions (pH 4), often implemented in microfluidic systems. Additionally, ionizable lipids offer the advantage of improving transfection efficiency, partly due to enhanced endosomal escape. Helper lipids, such as cholesterol, DSPC, or DOPC, contribute to the stability and rigidity of LNPs, and they also influence cellular processes such as endocytosis. The incorporation of PEGylated lipids into LNPs affects a number of physiological processes, including prolonged blood circulation, half-life, and in vivo distribution. Furthermore, these lipids influence the characteristics of LNPs, such as size, encapsulation efficiency, and aggregation.
The advantageous characteristics of lipids and their compositions in the formulation of RNA necessitate the utilization of suitable analytical methods throughout the drug development process. It is therefore essential that these methods enable the quantification of individual components and facilitate stability studies. To devise an analytical approach for lipid quantification, various chromatographic techniques can be explored. Among these, high-performance liquid chromatography (HPLC) stands out as a widely embraced method.
Different HPLC modes can be employed for analysis of lipids; normal phase liquid chromatography (NPLC) [7–9], and most commonly used reverse phase liquid chromatography (RPLC) [10]. In RPLC, a non-polar stationary phase (typically C18, C8) is used, and separation is based on the hydrophobic interactions between the lipid molecules and the stationary phase [11–13]. The choice of detector in HPLC hinges upon the properties of the analytes under examination. Given that most lipids lack chromophores, UV detectors may prove inadequate for analyzing LNPs. Other analytical techniques, such as mass spectrometry (MS) [14–16], refractive index detectors (RID) [17], evaporative light scattering detectors (ELSD) [18], and charged aerosol detector (CAD) [19–21] have been employed for lipid analysis. However, MS is often considered too costly for routine use, and developing a thoroughly validated method presents challenges. RID suffers from inherent limitations, primarily low sensitivity, while CAD struggles with poor signal-to-noise ratios. ELSD is garnering significant interest due to its capability to measure charged particle signals with an electrometer. Moreover, its response is generally independent of the chemical structure of the analyte, with volatility being a more critical factor.
In this study, a reverse phase liquid chromatography (RPLC) method, utilizing a PATfix® chromatographic system equipped with an ELSD and a monolith CIMac™ C4 HLD analytical column, was developed to separate and quantify lipids and nucleic acid in different LNP formulations. The developed analytical method was applied to analyze the lipid composition of LNPs and to compare the lipid components across different LNP formulations. This method allows for a direct injection of LNP formulations into the chromatographic system, obviating the need for dissolution or disassembly of the LNPs.
Materials and methods
Materials
Triethylammonium acetate buffer (TEAA, HPLC grade) and sucrose (≥99.5) were obtained from Sigma-Aldrich (Taufkirchen, Germany), Dulbecco’s phosphate buffered saline (PBS) from VWR (Lutterworth, UK) and isopropanol (IPA, LC-MS grade) from Merck (Darmstadt, Germany). Buffers were freshly prepared with LC-MS grade water purchased from Honeywell (Seelze, Germany). mFix4 was obtained from Sartorius BIA Separations (Ajdovščina, Slovenia), cationic lipid LipidBrick® IM21.7c from Polyplus (Illkirch, France), ionizable lipid SM-102 from Biosynth (Bratislava, Slovakia) and cholesterol from Sigma-Aldrich (Taufkirchen, Germany), while lipids DOPC, DSPC and DMG-PEG2k were purchased from Avanti (Alabaster, AL, USA).
LNP sample preparation
Two LNPs with different lipid composition were prepared. Lipids used for cationic lipid LNP were LipidBrick® IM21.7c, DOPC, cholesterol and DMG-PEG2k (molar ratio 33.3:6.7:25.7:1.0), while SM-102, DSPC, cholesterol and DMG-PEG2k (molar ratio 28.9:5.6:26.7:1.0) were used for ionizable lipid LNP. For both LNPs, mRNA mFix4 was used. LNPs were prepared using NanoAssemblr™ Ignite™ system with NxGen™ cartridge (Precision NanoSystems, Vancouver, Canada) and were buffer exchanged to storage buffer (1× PBS, 10% sucrose, pH 7.4) with 30 kDa cut-off Amicon filters (Merck, Darmstadt, Germany).
PATfix analytical system connected to ELSD
Chromatographic experiments were performed using PATfix analytical system, with a quaternary pump, a multiwavelength UV–Vis detector, a column thermostat and 8-port valve (Sartorius BIA Separations). For lipid detection, SEDEX LT-ELSD LC™ (Knauer, Berlin, Germany) was used. Samples were analyzed with 0.1 mL CIMac C4 analytical column (2 mL channel size) (Sartorius BIA Separations). Before analysis, samples were diluted with 10 mM TEAA and IPA (3:1). Injection volume was 500 mL. Sample analysis was monitored using UV detection at 260 and 280 nm. ELSD evaporation temperature was set to 53 °C and the C4 column was kept at 30 °C. For instrument control and data processing, PATfix software (Sartorius BIA Separations) was used.
Results and discussion
Lipids and mRNA separation and determination
The analysis of LNPs in a single assay presents a significant challenge due to their complex composition, which typically includes nucleic acid and at least four different lipid species. The diverse polar properties of these components make comprehensive analysis on reverse-phase columns difficult. A challenge arises from the suboptimal interaction between polar nucleic acids and the polar head groups on lipids with reverse-phase columns [18]. Consequently, the employment of an alternative column or the modification of the mobile phase may result in enhanced outcomes. Zhong and colleagues initially proposed an effective approach for the detection and separation of DOTAP using a C18 column, with TFA included in the mobile phase. TFA serves to protect the positively charged headgroups of cationic and ionizable lipids, thereby extending their interaction with the stationary phase.
The objective of this study is to expand the scope of lipid analysis assays to encompass the detection of nucleic acid in LNP formulations. The substitution of TFA with the less aggressive ion-pairing reagent TEAA played a pivotal role in alleviating the issue of excessive and irreversible interaction between mRNA in LNP formulations and the C4 reverse-phase column. This substitution effectively facilitated the separation of all lipids in the LNP formulation, while promoting efficient binding of the nucleic acid.
Figure 1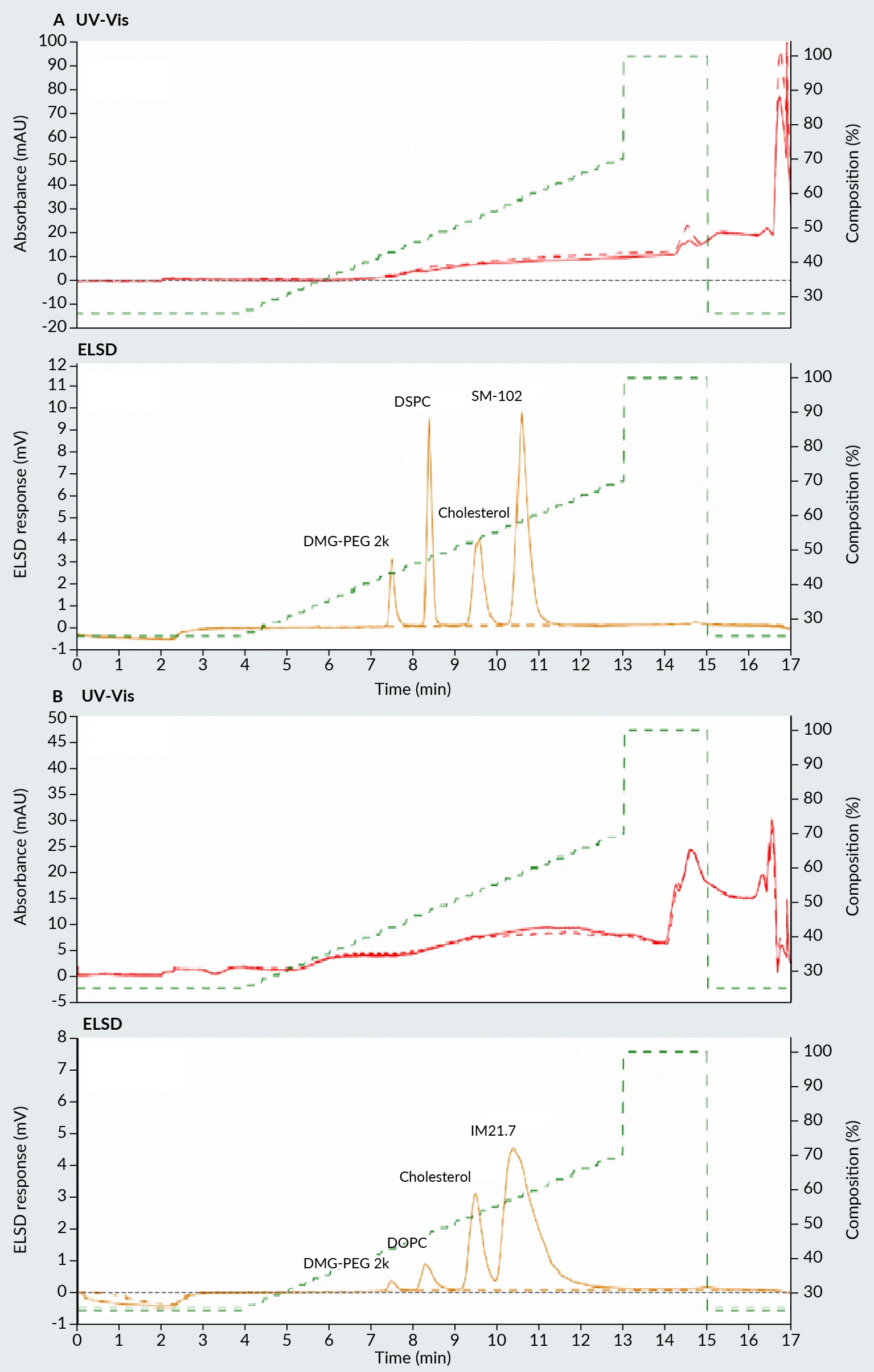 Chromatogram of four lipids used in preparation of LNP with (A) ionizable lipid, and (B) cationic lipid. Upper part of the chromatogram represents UV-Vis detector signal and lower part ELSD signal. LNP: Lipid nanoparticles; ELSD: Evaporative light scattering detectors. illustrates two examples of the separation of four distinct lipids commonly used in LNP formation. The upper section of the figure displays UV signals at 260 nm, while the lower section shows the ELSD signal trace. Due to the absence of chromophores, lipids do not exhibit any response in the UV section of the chromatogram. Instead, their presence is detected solely in the ELSD section. In Figure 1A, an ionizable lipid is employed alongside cholesterol, a phospholipid, and a pegylated lipid. In Figure 1B, a cationic lipid is used in conjunction with cholesterol, a phospholipid, and a pegylated lipid. In both scenarios, separation was achieved using a C4 HLD column with ELSD detection in an increased isopropanol gradient. The method yielded distinct peaks and a baseline separation of all four lipids. The elution order of all four lipids is governed by their hydrophobic nature. The more polar lipids are eluted first, followed by the phospholipid, cholesterol, and then the ionizable/cationic lipid, respectively.
Chromatogram of four lipids used in preparation of LNP with (A) ionizable lipid, and (B) cationic lipid. Upper part of the chromatogram represents UV-Vis detector signal and lower part ELSD signal. LNP: Lipid nanoparticles; ELSD: Evaporative light scattering detectors. illustrates two examples of the separation of four distinct lipids commonly used in LNP formation. The upper section of the figure displays UV signals at 260 nm, while the lower section shows the ELSD signal trace. Due to the absence of chromophores, lipids do not exhibit any response in the UV section of the chromatogram. Instead, their presence is detected solely in the ELSD section. In Figure 1A, an ionizable lipid is employed alongside cholesterol, a phospholipid, and a pegylated lipid. In Figure 1B, a cationic lipid is used in conjunction with cholesterol, a phospholipid, and a pegylated lipid. In both scenarios, separation was achieved using a C4 HLD column with ELSD detection in an increased isopropanol gradient. The method yielded distinct peaks and a baseline separation of all four lipids. The elution order of all four lipids is governed by their hydrophobic nature. The more polar lipids are eluted first, followed by the phospholipid, cholesterol, and then the ionizable/cationic lipid, respectively.
In order to assess the suitability of the method for LNP formulation investigation, LNPs were directly analyzed using this method without any sample pre-treatment, with the exception of dilution with the loading buffer. The resulting chromatograms of LNPs containing ionizable and cationic lipid are shown in Figure 2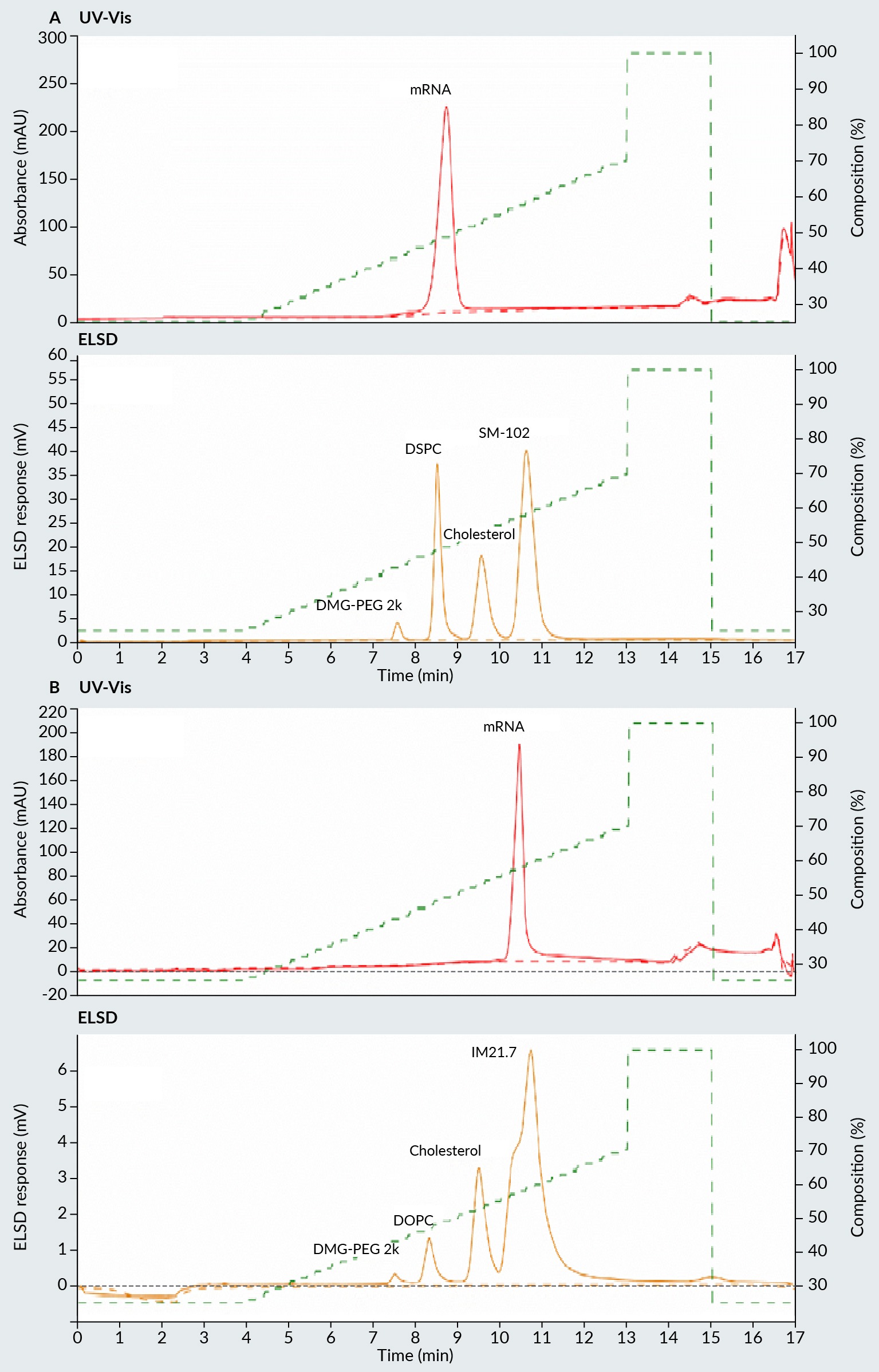 Chromatogram of two LNP samples prepared with (A) ionizable lipid, and (B) cationic lipid.Upper part of the chromatogram represents UV-Vis detector signal and lower part ELSD signal. A & B, respectively.
Chromatogram of two LNP samples prepared with (A) ionizable lipid, and (B) cationic lipid.Upper part of the chromatogram represents UV-Vis detector signal and lower part ELSD signal. A & B, respectively.
In the case of the ionizable lipid LNP formulation (Figure 2A), it can be demonstrated that all the lipids composing the LNPs are effectively separated. Furthermore, an additional peak is observed in the UV region of the chromatogram, which represents the mRNA. It is crucial to acknowledge that this analytical method is unable to differentiate between naked mRNA and encapsulated mRNA. The LNPs were destroyed on the C4 column during the analysis, resulting in the co-elution of naked and encapsulated mRNA. A comparable observation is made for cationic lipid LNPs (Figure 2B), where all four lipids are distinctly separated in the ELSD portion of the chromatogram. In this instance, the mRNA peak is also observed in the UV spectra, indicating the elution of the mRNA. For cationic lipid LNPs, the mRNA elutes at a higher concentration of isopropanol, indicating greater hydrophobicity of the eluted mRNA. This phenomenon may result from the presence of the permanently charged cationic lipid, which can act as an ion-pairing reagent. The polar, positively charged head of the cationic lipid interacts with the negatively charged phosphate groups on the mRNA, while the non-polar tail of the lipid strongly interacts with the C4 column, thus prolonging the elution of mRNA. This interaction illustrates the reason why the elution of the mRNA occurs simultaneously with the elution of the cationic lipid from the C4 column. Consequently, the ELSD elution peak of the cationic lipid exhibits a different shape compared to the elution profile when mRNA is not present in the sample. In addition to the ability to detect and quantify the lipids, this method also permits the detection and quantification of the total mRNA in LNPs in the same assay.
Method validation
The developed chromatographic method has been validated by evaluating several criteria, including sensitivity (limits of detection and quantification), linearity, precision (repeatability, intermediate repeatability) and accuracy all included in ICH guidance for analytical method validation [22]. The validation results are presented in Table 1. LOD and LOQ for mRNA were determined from signal to noise ratio of 3 and 10, respectively, using UV 260 nm absorbance signal. For LOD and LOQ of lipids, ELSD signal was used instead (Table 1). The linearity of the method was evaluated at six concentration levels for each of the six lipids and mRNA. ELSD signal does not vary linearly as function of the injected mass but in our case follows polynomial model (ax2 + bx + c) (Table 1). In the case of UV detection of mRNA, normal linear model was used (y=kx + n) (Table 1). The precision of the method was evaluated by analyzing standard mixtures of the four lipids composing LNP sample with ionizable lipid the at 1 µg/mL each in ethanol. The relative standard deviation (RSD) values of the peak areas measured by ELSD from six consecutive injections of the same standard lipid mixture were calculated to check the method’s repeatability (Table 1). In order to assess the accuracy of the method, mixtures of the four lipids composing LNP sample with ionizable lipid were prepared at known concentrations of 1 mg/mL in ethanol. The recoveries, expressed as a percentage, between the known concentrations and the calculated concentrations of lipids, based on the calibration curves, were determined (Table 1).
| Table 1. Validation results. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Retention time (min) | Calibration curve | R2 mRNA | LOD (µg) mRNA | LOQ (µg) mRNA | RSD (peak areas; %) n = 6 | Recoveries (%) n = 6 (ELSD) | Recoveries RSD (%) n = 6 (ELSD) |
| mRNA | 9.1 | y=2602× − 56 | 0.9999 | 0.01 | 0.07 | N/A | N/A | N/A |
| IM21.7c | 10.4 | y=0.7171×2 + 4.7673× − 3.1714 | 0.9997 | 0.56 | 0.75 | N/A | N/A | N/A |
| DOPC | 8.2 | y=1.2365×2 + 10.886× − 8.8627 | 0.9989 | 0.57 | 0.60 | N/A | N/A | N/A |
| SM-102 | 10.6 | y=1.423×2 + 5.5894x − 1.9832 | 0.9994 | 0.44 | 0.55 | 1.0 | 91.0 | 0.5 |
| DSPC | 8.4 | y=0.9497×2 + 28.134x − 28.049 | 0.9998 | 0.62 | 0.69 | 0.8 | 95.1 | 0.3 |
| Cholesterol | 9.5 | y=2.3862×2 + 18.396x − 23.192 | 0.9991 | 0.56 | 0.62 | 1.7 | 94.5 | 0.8 |
| DMG-PEG2k | 7.5 | y=1.0009×2 + 13.824x − 15.694 | 0.9991 | 0.54 | 0.58 | 1.2 | 91.6 | 0.5 |
Method application for LNP composition assessment
The applicability of the validated chromatographic method for lipid and mRNA quantification was evaluated by analyzing six different LNPs formulations. Three LNPs were prepared with ionizable lipids, and three with cationic lipids with addition of phospholipid, cholesterol and pegylated lipid. In addition to the different lipid compositions, the lipid ratios varied among the formulations. Samples of LNPs were obtained at various stages of preparation, including the lipid mix injected into the microfluidics machine, the crude LNP sample directly from the microfluidics machine, and the purified LNP sample obtained through tangential flow filtration (TFF). The nature and proportion of lipids must be tailored to each application, necessitating the quantitative analysis of each lipid [23]. Subsequently, the chromatographic method was employed as a quality control tool to monitor changes in lipid ratios of nanoparticles throughout the manufacturing process. Prior to subjecting the lipids to the microfluidic machine, the recoveries (percentage of each lipid compared to the theoretical quantity) were calculated for all lipids, with values ranging from 92.1%–107.1% for LNPs with ionizable lipid and from 96.7%–106.0%. This demonstrated the reliability of the developed chromatographic method (Figure 3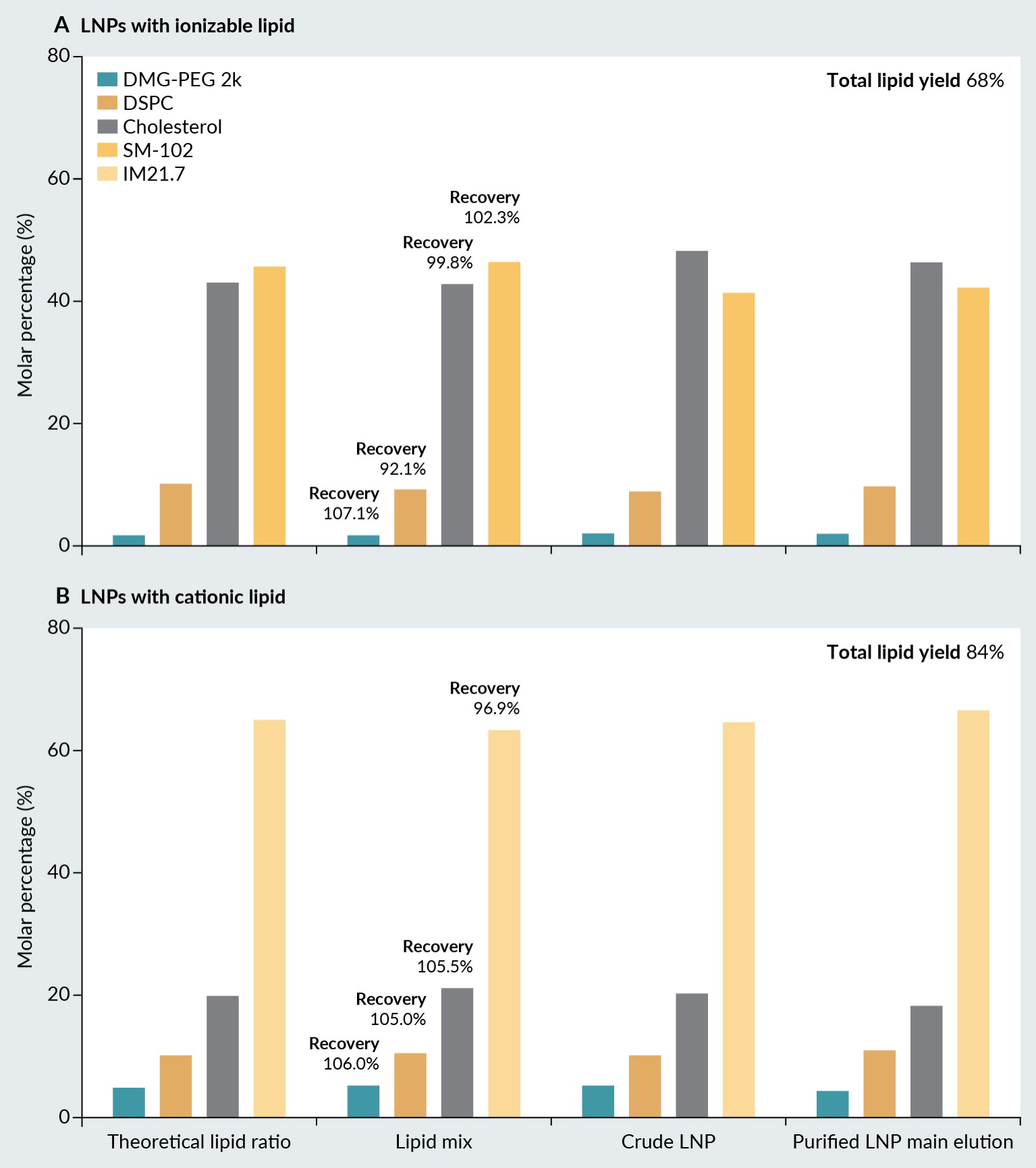 Molar percentage of the LNP building block lipids in LNP with (A) ionizable lipid, and (B) cationic lipid.).
Molar percentage of the LNP building block lipids in LNP with (A) ionizable lipid, and (B) cationic lipid.).
In the case of LNPs prepared with ionizable lipid, the molar percentage of ionizable lipid (SM-102) decreased from 46.5%–41.2% during microfluidic mixing. In contrast, the molar percentage of cholesterol increased from 42.9%–48.2%. The molar percentages of phospholipid and pegylated lipid remain unchanged during microfluidic mixing. TFF filtration does not affect the lipid ratio of the LNPs. The manufacturing process of the LNPs resulted in a loss of between 32% of the total lipid concentration (Figure 3), without inducing significant changes in lipid molar ratios. This loss of lipids was probably due to the elimination of lipids not involved in the lipid nanoparticles, which were able to pass through the TFF membrane.
A different observation was made with LNPs prepared with cationic lipid. In this case, the molar percentage of cationic lipid increased slightly from 63.0%–66.5% during microfluidic mixing and TFF filtration. Consequently, a slight decrease in the molar percentage of cholesterol is observed. Again, the molar percentage of phospholipid and pegylated lipid remained the same. The loss of lipids was not as pronounced as in the case of ionizable lipids, only about 16% of the total lipid concentration was observed (Figure 3).
Conclusion
This study presents the development and validation of a novel reverse phase liquid chromatography method for the quantitative analysis of lipids and nucleic acids in LNPs. The method, which utilizes an evaporative light scattering detector and CIMac C4 HLD monolithic column, enables direct injection of LNP formulations, allowing simultaneous separation and quantification of both lipid components and nucleic acid without the need for sample pre-treatment. The utilization of a monolith CIMac C4 HLD column enabled the developed method to achieve effective separation and distinct detection of all lipid constituents. The method was demonstrated to be highly sensitive, linear, precise, and accurate, meeting the specifications included in ICH guidelines, thereby proving its reliability for routine analytical applications. Furthermore, the method revealed its ability to track changes in lipid composition throughout the LNP manufacturing process, thereby ensuring the stability and consistency of LNP formulations.
The application of the method to a range of LNP formulations, including both ionizable and cationic lipids, demonstrated its robustness in detecting differences in lipid ratios and mRNA. This capability highlights the potential of the method as a critical quality control tool, aiding in the reproducibility and efficacy of LNP-based therapeutics. Consequently, this advanced analytical approach supports the ongoing development and optimization of LNPs for diverse therapeutic applications, potentially leading to significantly improved LNP-based therapeutics.
References
- Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021; 6(12), 1078–1094. Crossref
- Li Y, Ye Z, Yang H, Xu Q. Tailoring combinatorial lipid nanoparticles for intracellular delivery of nucleic acids, proteins, and drugs. Acta Pharm. Sin. B 2022; 12(6), 2624–2639. Crossref
- Schlich M, Palomba R, Costabile G, et al. Cytosolic delivery of nucleic acids: the case of ionizable lipid nanoparticles. Bioeng. Transl. Med. 2021; 6, 2. Crossref
- Li M, Li Y, Li S, et al. The nano delivery systems and applications of mRNA. Eur. J. Med. Chem. 2022; 227, 113910. Crossref
- Kulkarni JA, Darjuan MM, Mercer JE, et al. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano 2018; 12(5), 4787–4795. Crossref
- Hald Albertsen C, Kulkarni JA, Witzigmann D, Lind M, Petersson K, Simonsen JB. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022; 188, 114416. Crossref
- McLaren DG, Miller PL, Lassman ME, Castro-Perez JM, Hubbard BK, Roddy TP. An ultraperformance liquid chromatography method for the normal-phase separation of lipids. Anal. Biochem. 2011; 414(2), 266–272. Crossref
- Abreu S, Solgadi A, Chaminade P. Optimization of normal phase chromatographic conditions for lipid analysis and comparison of associated detection techniques. J. Chromatogr. A 2017; 1514, 54–71. Crossref
- Hutchins PM, Barkley RM, Murphy RC. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 2008; 49(4), 804–813. Crossref
- Peterson BL, Cummings BS. A review of chromatographic methods for the assessment of phospholipids in biological samples. Biomed. Chromatogr. 2006; 20(3), 227–243. Crossref
- Lin JT, Mckeon TA, Woodruff CL, Singleton JA. Separation of synthetic phosphatidylcholine molecular species by high-performance liquid chromatography on a C column. J. Chromatogr. A 1998; 824(2), 169–174. Crossref
- Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem. 2014; 61, 192–206. Crossref
- Suchocka Z, Gronostajska D, Suchocki P, Pachecka J. New HPLC method for separation of blood plasma phospholipids. J. Pharm. Biomed. Anal. 2003, 32, 859–865. Crossref
- Knittelfelder OL, Weberhofer BP, Eichmann TO, Kohlwein SD, Rechberger GN. A versatile ultra-high performance LC-MS method for lipid profiling. J. Chromatogr. B 2014; 1, 119–128. Crossref
- Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011; 80(1), 301–325. Crossref
- Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem. 2014; 61, 192–206. Crossref
- Grit M, Crommelin DJA, Lang J. Determination of phosphatidylcholine, phosphatidylglycerol and their lyso forms from liposome dispersions by high-performance liquid chromatography using high-sensitivity refractive index detection. J. Chromatogr. A 1991; 585(2), 239–246. Crossref
- Zhong Z, Ji Q, Zhang JA. Analysis of cationic liposomes by reversed-phase HPLC with evaporative light-scattering detection. J. Pharm. Biomed. Anal. 2010; 51(4), 947–951. Crossref
- Bender V, Fuchs L, Süss R. RP-HPLC-CAD method for the rapid analysis of lipids used in lipid nanoparticles derived from dual centrifugation. Int. J. Pharm. X 2024; 7, 100255. Crossref
- Kim KH, Lee JE, Lee JC, et al. Optimization of HPLC CAD method for simultaneous analysis of different lipids in lipid nanoparticles with analytical QbD. J. Chromatogr. A. 2023; 1709, 464375. Crossref
- Yu X, Yu C, Wu X, et al. Validation of an HPLC-CAD method for determination of lipid content in LNP-encapsulated COVID-19 mRNA vaccines. Vaccines (Basel) 2023; 11(5), 937. Crossref
- EMA. ICH Q2(R2) Guideline on Validation of Analytical Procedures. 2024; European Medicines Agency. https://www.ema.europa.eu/en/ich-q2r2-validation-analytical-procedures-scientific-guideline. Crossref
- Eygeris Y, Gupta M, Kim J, Sahay G. Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 2022; 55(1), 2–12. Crossref
Affiliations
Nejc Pavlin
Sartorius BIA Separations,
Ajdovščina, Slovenia
Mojca Bavčar
Sartorius BIA Separations,
Ajdovščina, Slovenia
Tristan Kovačič
Sartorius BIA Separations,
Ajdovščina, Slovenia
Tjaša Plesničar
Sartorius BIA Separations,
Ajdovščina, Slovenia
Andreja Gramc Livk
Sartorius BIA Separations,
Ajdovščina, Slovenia
Aleš Štrancar
Sartorius BIA Separations,
Ajdovščina, Slovenia
Authorship & Conflict of Interest
Contributions: The named authors take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Acknowledgements: None.
Disclosure and potential conflicts of interest: The authors are employees of Sartorius BIA Separations d.o.o.
Funding declaration: The authors received no financial support for the research, authorship and/or publication of this article.
Article & Copyright Information
Copyright: Published by Cell & Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0 which allows anyone to copy, distribute, and transmit the article provided it is properly attributed in the manner specified below. No commercial use without permission.
Attribution: Copyright © 2024 Sartorius BIA Separations. Published by Cell & Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0.
Article source: Invited; externally peer reviewed.
Submitted for peer review: Jun 21, 2024; Revised manuscript received: Jul 9, 2024; Publication date: Jul 30, 2024.

